Перекрёстные ссылки книги для 1266
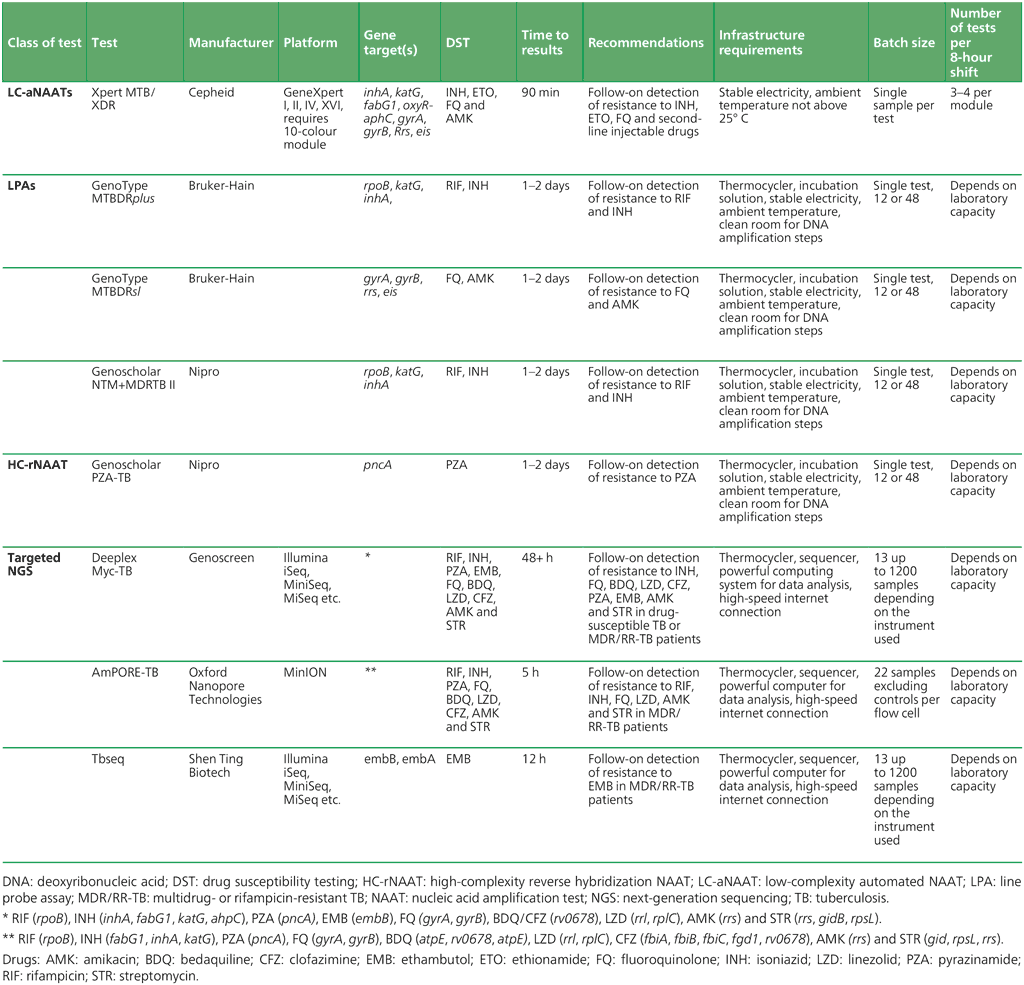
The availability of follow-on tests to detect additional resistance to anti-TB drugs is becoming increasingly important for making informed decisions on treatment regimens. WHO has recommended four classes: LC-aNAAT for detection of resistance to INH and second-line anti-TB drugs, LPAs, high-complexity reverse hybridization NAATs (HC-rhNAATs) and targeted NGS. A summary of the tests included in the different classes is shown in Table 2.6.
Table 2.6. Comparison of follow-on tests for the detection of drug resistance
2.4.1 LC-aNAATs for detection of resistance to INH and second-line anti-TB drugs
The first-in-class product for LC-aNAATs for detection of resistance to INH and second-line anti-TB drugs was the Xpert MTB/XDR Assay (Cepheid, Sunnyvale, USA); the class is defined in the consolidated guidelines (5) and in Table 2.3. This test uses a cartridge designed for GeneXpert instruments to detect resistance to INH, FQs, ETO and second-line injectable drugs (e.g. AMK). In contrast to Xpert MTB/RIF and Xpert Ultra, which are performed on a GeneXpert instrument that can detect six colours, the XDR test requires a 10-colour GeneXpert module. There is an option to combine the 6- and 10-colour systems through a common computer, or to replace one 6-colour module in an instrument with a 10-colour module. The current WHO recommendations for Xpert Ultra cartridge use on GeneXpert 6-colour instruments are also valid for their use on GeneXpert 10-colour instruments (21) (Box 2.5).

The LC-aNAAT is intended for use as a follow-on test in specimens determined to be MTBC-positive; it offers the chance to improve access to rapid DST in peripheral laboratories. In addition, the test provides results in less than 90 minutes, meaning that the time to results is faster than with LPAs or culture-based phenotypic DST. This NAAT requires the same infrastructure and training of technicians as the other Xpert tests.
The overall pooled sensitivity for detection of INH resistance was 94% (95% CI: 89–97%) and specificity was 98% (95% CI: 95–99%) (Table 3.3). Overall pooled sensitivity for detection of FQ resistance was 93% (95% CI: 88–96%) and specificity was 98% (95% CI: 94–99%) (Table 3.4). Thus, Xpert MTB/XDR could be used as a reflex test to complement existing technologies that only test for RIF resistance, allowing the rapid and accurate detection of resistance to INH and FQs in cases of RIF-susceptible TB; and of resistance to FQ, INH, ETO and AMK in cases of RR-TB. The package insert includes the use of the test on culture isolates. However, the primary purpose of this test is to achieve rapid and early detection of resistance, and recommendations are for use of the test directly on clinical specimens. Web Annex A includes an information sheet summarizing the procedure, and providing operational and implementation considerations for this test.
2.4.2 Line probe assays
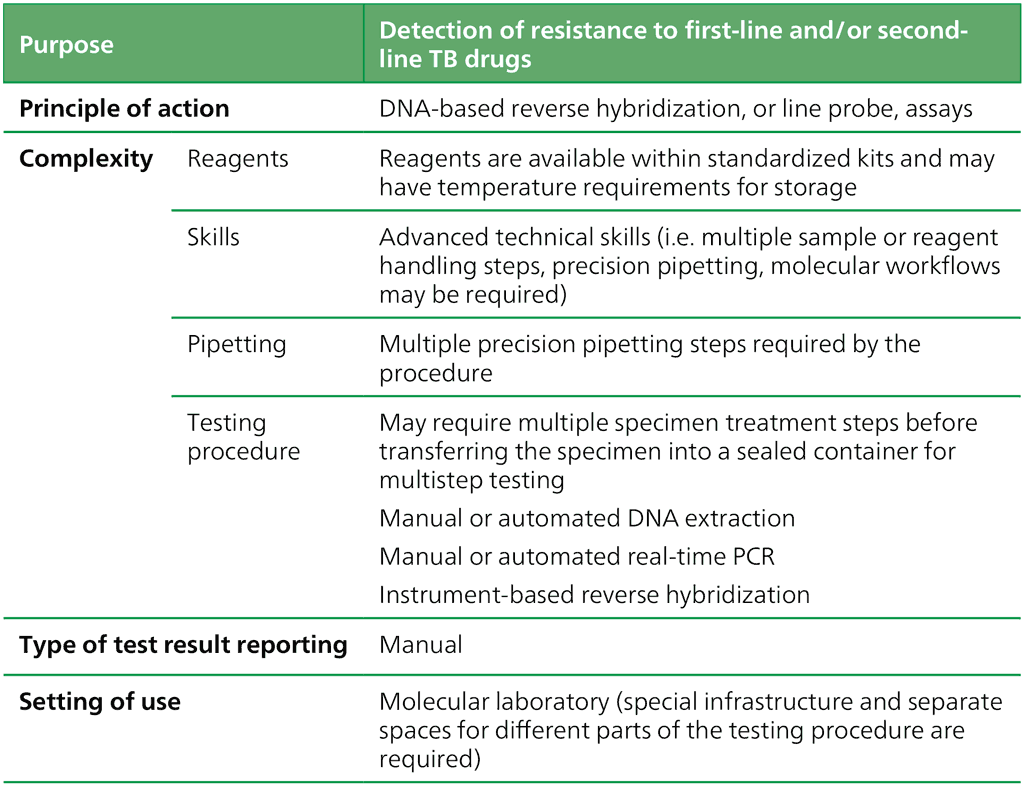
LPAs are a family of hybridization tests based on DNA strips that detect the presence or absence of mutations associated with drug resistance. The assays do this directly, through binding DNA amplification products (amplicons) to probes targeting the most commonly occurring mutations (MUT probes), or indirectly, by inferring resistance through the binding of a wildtype probe to a wild-type target sequence. The definition of the class is found in Table 2.7.
Table 2.7. Definition of the class criteria for LPAs
First-line drug LPAs
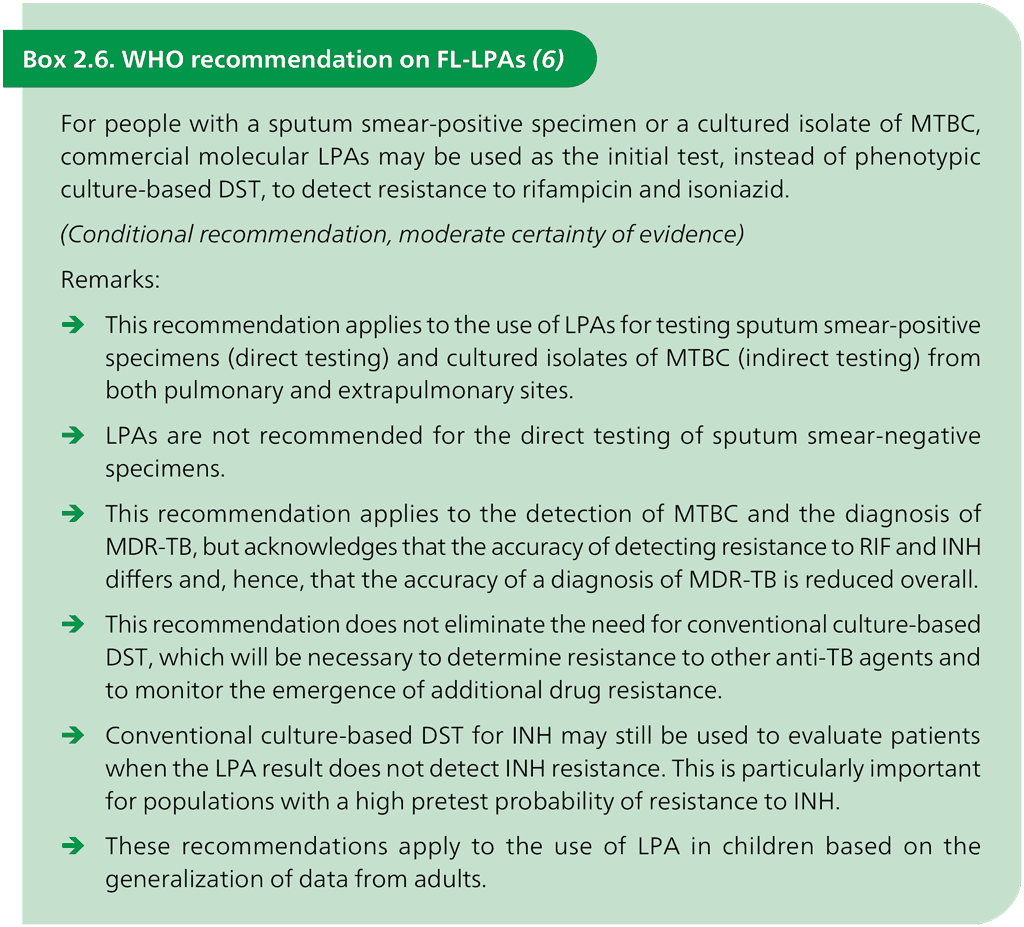
First-line drug LPAs (FL-LPAs) such as the GenoType MTBDRplus test and the NTM+MDRTB Detection Kit allow the detection of resistance to RIF and INH. WHO recommends using FL-LPAs in the situations shown in Box 2.6.
Second-line drug LPAs
Second-line drug LPAs (SL-LPAs) such as the GenoType MTBDRsl test allow the detection of resistance to FQs and AMK (Box 2.7).

2.4.3 High-complexity reverse hybridization NAATs
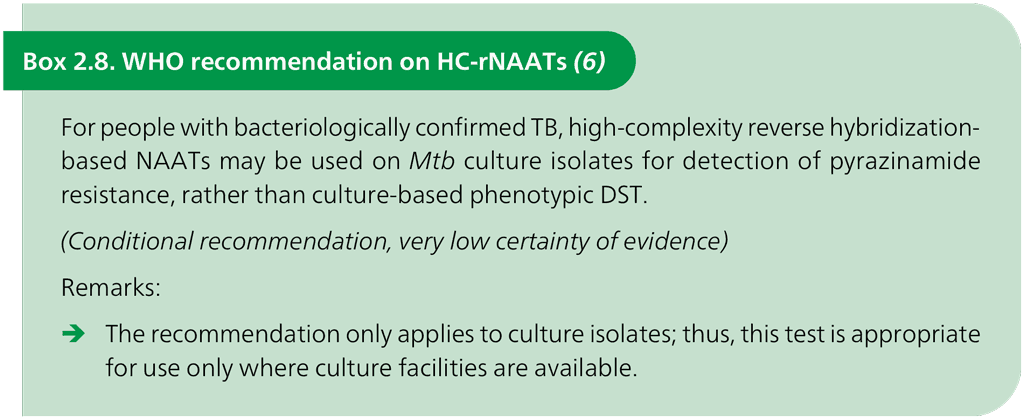
The first-in-class product for the class of HC-rNAATs is the Genoscholar PZA-TB (Nipro, Osaka, Japan) for the detection of resistance to PZA. The Genoscholar PZA-TB test is based on the same principle as the FL-LPA and SL-LPA but requires the use of many hybridization probes to cover the full pncA gene (>700 base pairs [bp]). Reading the hybridization results on the strips with a total of 48 probes requires careful attention; however, the test provides faster results than phenotypic DST and is based on molecular detection. The overall pooled sensitivity for the detection of PZA resistance was 81.2% (95% CI: 75.4–85.8%) and the pooled specificity was 97.8% (95% CI: 96.5–98.6%) (22). The hybridization can be performed on the TwinCubator instruments (Bruker-Hain, Germany) that are used for LPAs (19). An information sheet summarizing HC-rNAATs is available in Web Annex A. The WHO recommendation on HC-rNAATs is given in Box 2.8.
2.4.4 Targeted NGS end-to-end solutions
The class of targeted NGS tests was defined as workflows that use massively parallel sequencing to detect resistance to TB drugs, starting from a processed clinical sample and concluding with an end-user report that relates detected Mtb mutations to the presence (or absence) of drug resistance, based on the interpretation of the WHO catalogue of mutations (23). Solutions that involve end-to-end targeted NGS tests couple DNA extraction and amplification of selected genes with NGS technology to detect resistance to many drugs with a single test, as shown in Fig. 2.2. Because targeted NGS tests can interrogate entire genes to identify specific mutations associated with resistance, these tests may provide improved accuracy compared with other WRDs. In addition, targeted NGS tests can detect resistance to new and repurposed drugs not currently included in any other molecular assays. Thus, these tests offer great potential to provide comprehensive resistance detection matched to modern treatment regimens (24).
Fig. 2.2. Process of testing, from sample to result report, for targeted NGS
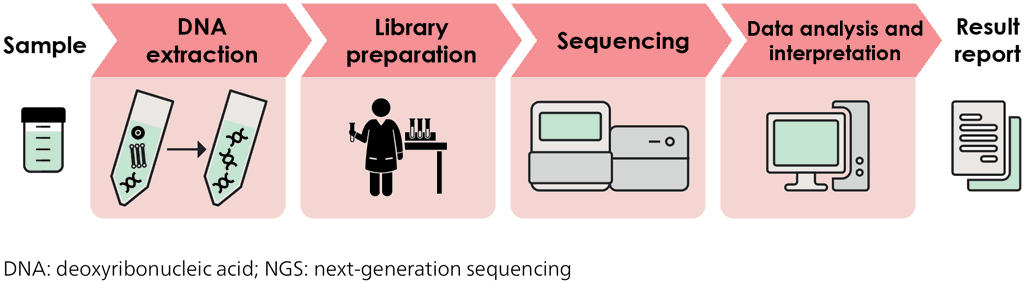
In 2023, WHO recommended the use of the class of targeted NGS to detect drug resistance for clinical decision-making, and included three solutions as first-in-class technologies. In January 2025, the WHO TAG assessed new and updated targeted NGS solutions that met class criteria. Based on the evidence available and outcomes of the evidence assessment, no new technologies were added to the targeted NGS class. However, the resistance interpretation software for the AmPORE-TB solution had been updated, and this product was found to meet class-based performance criteria for detection of resistance to three additional drugs (PZA, BDQ and CFZ). The current list of targeted NGS solutions and the drugs for which WHO recommends their use for resistance detection are given in Table 2.8.
Table 2.8. List of targeted NGS solutions and drugs for which WHO recommends their use
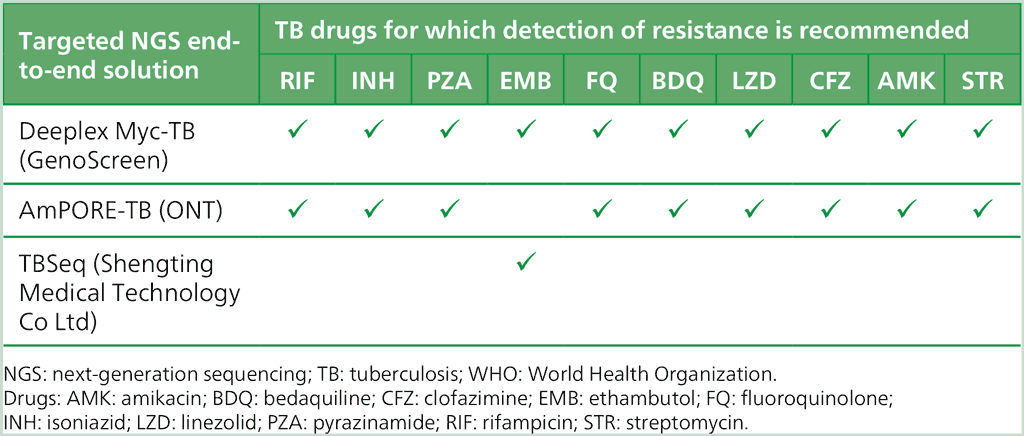
The WHO policy statement on the outcomes of the TAG meeting is summarized in Box 2.9. Further considerations for the implementation of targeted NGS solutions are outlined in Annex 3; these considerations are supported by the WHO manual on the use of NGS for the surveillance of DR-TB (25).

NEW
The TAG report outlines plans for how targeted NGS diagnostic class performance will be updated as individual product performance improves over time.
WHO provides two recommendations on the use of targeted NGS (Box 2.10). Among people with bacteriologically confirmed pulmonary TB, the targeted NGS test performances (Table 3.3, Table 3.4 and Table 3.5 in Section 3) were determined to be accurate for all drugs included in the assessment, with pooled sensitivity of at least 95% for INH, MFX and ethambutol (EMB); more than 93% for RIF and levofloxacin (LFX); and 88% for PZA.⁴ The pooled specificity was at least 96% for all drugs. The reference standard was phenotypic DST for INH, LFX and MFX, and a combination of phenotypic DST and whole-genome sequencing (WGS) for RIF, PZA and EMB. The indeterminate rate ranged from 7% (AMK) to 18% (PZA) but was highest in samples with low or very low bacterial loads, which may have implications for implementation; therefore, priority should be given to samples with a higher bacillary load. The overall certainty of the evidence ranged from low to moderate for test accuracy.
Among people with bacteriologically confirmed RIF-resistant pulmonary TB, the targeted NGS test performances (Table 3.6 in Section 3) were determined to be accurate for INH, LFX, MFX, streptomycin (STR) and EMB (pooled sensitivity ≥95%), and acceptable for BDQ (68%), LZD (69%), CFZ (70%), AMK (87%) and PZA (90%). The specificity was at least 95% for all drugs except STR (75%). The reference standard was phenotypic DST for all drugs except EMB and PZA, where a combination of phenotypic DST and WGS was used. The indeterminate rate ranged from 9% (LFX and MFX) to 21% (EMB) and depended on the bacterial load. For test accuracy, the overall certainty of the evidence ranged from low to high.

All products recommended by WHO are automatically eligible to be included in the WHO essential diagnostic list. The WHO recommendations on diagnostics are based on clinical research evidence; they do not include quality assessments of the products or the manufacturing process involved. Before introducing any new products, countries should ensure that those products fulfil local or internationally recognized regulatory requirements.
4 Pooled sensitivity and specificity for targeted NGS were calculated on the first-in-class tests when the class recommendation was made; values may be updated in the future if expected improvements in performance eventuate. Considerations on performance value updates for the targeted NGS class can be found in the TAG meeting report in Web Annex D.

 Обратная связь
Обратная связь