Book traversal links for 1266
2.4.1 Low complexity automated NAATs for detection of resistance to isoniazid and second-line anti-TB agents
Among 105 countries possessing representative data on resistance to fluoroquinolones from the past 15 years, the proportion of MDR/RR-TB cases with resistance to any fluoroquinolone for which testing was done was 20.1% (95% CI: 15.5–25.0%) (1). Thus, rapid and early testing for the detection of fluoroquinolone resistance is essential for determining eligibility for treatment with the all-oral 9–12 month standardized shorter regimen for MDR/RR-TB. However, the current limitation with testing for fluoroquinolone resistance is the limited accessibility of current technologies (which are often only available at higher tiers of the health system) and poor yield in paucibacillary specimens.
Low complexity automated NAATs are a new class of diagnostics intended for use as a reflex test in specimens determined to be Mtb complex (MTBC)-positive; they offer rapid DST in intermediate and peripheral laboratories. The first product in this class simultaneously detects resistance to isoniazid, fluoroquinolones, ethionamide and amikacin. Results are available in under 90 minutes, leading to faster time to results than the current standard of care, which includes LPAs and culture-based phenotypic DST.
An additional value of the tests is the accurate and rapid detection of isoniazid resistance, which is relevant for both RR-TB and rifampicin-susceptible TB; the latter is often undiagnosed and contributes to a large burden of disease. Globally, rifampicin-susceptible TB is estimated to occur in 13.1% (95% CI: 9.9–16.9%) of new cases and 17.4% (95% CI: 0.5–54.0%) of previously treated cases (1). Thus, this test could also be used as a reflex test to complement existing technologies that only test for rifampicin, allowing the rapid and accurate detection of isoniazid-resistant, rifampicin-susceptible TB.
Although these new technologies are excellent at detecting resistance to selected drugs, conventional culture-based phenotypic DST remains important to determine resistance to other anti-TB agents, particularly the new and repurposed medicines such as bedaquiline and linezolid.
Recommendations



There are several subgroups to be considered for these recommendations:
- The recommendations are based on the evidence of diagnostic accuracy in sputum of adults with bacteriologically confirmed pulmonary TB, with or without rifampicin resistance.
- The recommendations are extrapolated to adolescents and children, based on the generalization of data from adults.
- The recommendations apply to people living with HIV (studies included a varying proportion of such individuals); data stratified by HIV status were not available.
- The recommendations are extrapolated to people with extrapulmonary TB, and testing of non-sputum samples was considered appropriate, which affects the certainty. The panel did not evaluate test accuracy in non-sputum samples directly, including in children; however, extrapolation was considered appropriate given that WHO has recommendations for similar technologies for use on non-sputum samples (e.g. Xpert MTB/RIF and Xpert Ultra).
- Recommendations for detection of resistance to amikacin and ethionamide are only relevant for people who have bacteriologically confirmed pulmonary TB and resistance to rifampicin.
Justification and evidence
The WHO Global TB Programme initiated an update of the current guidelines and commissioned a systematic review on the use of low complexity automated NAATs for the detection of resistance to isoniazid and second-line TB drugs in people with signs and symptoms of TB.
The PICO questions were designed to form the basis for the evidence search, retrieval and analysis:
- Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, irrespective of resistance to rifampicin, for detection of resistance to isoniazid, as compared with culture-based phenotypic DST?
- Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, irrespective of resistance to rifampicin, for detection of resistance to fluoroquinolones, as compared with culture-based phenotypic DST?
- Should low complexity automated NAATs be used on culture isolates in people with signs and symptoms of pulmonary TB, and detected resistance to rifampicin, for detection of resistance to ethionamide, as compared with genotypic sequencing of the inhA promoter?
- Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, and detected resistance to rifampicin, for detection of resistance to amikacin, as compared with culture-based phenotypic DST?
The databases Ovid Medline (Ovid, 1946 to present) and Embase (Ovid, 1947 to present) were searched for studies evaluating cartridge-based tests using the following search terms: tuberculosis, pulmonary AND Xpert, GeneXpert, Truenat, cartridge, point-of-care systems, drug susceptibility test, isoniazid resistance, fluoroquinolone resistance and second-line injectable drug resistance. Clinicaltrials.gov and the WHO International Clinical Trials Registry Platform were also searched for trials in progress. Searches were run up to 6 September 2020 without language restriction. On 4 November 2020, an additional search was run using the search terms Zeesan and MeltPro.
Researchers at FIND, the WHO Global TB Programme, the manufacturer and other experts in the field of TB diagnostics were contacted for information about ongoing and unpublished studies. Data submitted in response to the WHO public call were reviewed.
Drug resistance was compared against a phenotypic reference standard (or a genotypic reference standard for ethionamide resistance), as well as a composite reference standard that was constructed by combining the results of phenotypic and genotypic DST results in studies where both had been performed.
Data synthesis was structured around the four preset PICO questions, as outlined below. Three web annexes give additional information, as follows:
- details of studies included in the current analysis (Web Annex A.6: Low complexity automated NAATs);
- a summary of the results and details of the evidence quality assessment (Web Annex A.6: Low complexity automated NAATs); and
- a summary of the GDG panel judgements (Web Annex A.6: Low complexity automated NAATs).
PICO 1: Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, irrespective of resistance to rifampicin, for detection of resistance to isoniazid, as compared with culture-based phenotypic DST?
Three multinational studies with 1605 participants provided data for evaluating isoniazid resistance detection. The reference standard for each of these studies was culture-based phenotypic DST. Each study centre in the multinational studies was analysed as a separate study (Fig. 2.4.1.1).
Several concerns were expressed about indirectness in the study populations. First, the median prevalence of isoniazid resistance in the included studies was 67.2% (range, 26.8% [Diagnostics for Multidrug Resistant Tuberculosis in Africa – DIAMA, Benin] to 93.9% [FIND, Moldova]), which is higher than the global estimates for isoniazid resistance. Hence, applicability to settings with a lower prevalence of isoniazid resistance comes with some uncertainty. Second, there are potential differences in the mutations present in isoniazid monoresistant strains and MDR strains; that is, some studies suggest that the mutations found in monoresistant strains are more diverse than the mutations found in MDR strains. Third, although the population for this PICO question is “irrespective of rifampicin resistance”, enrolment criteria in the studies meant that most participants within the included studies had RR-TB. As a result of these concerns, certainty of evidence was downgraded one level for indirectness both for sensitivity and specificity, and the quality (certainty) of evidence was rated moderate both for sensitivity and specificity.
Fig. 2.4.1.1 Forest plot of included studies for isoniazid resistance detection, irrespective of rifampicin resistance with culture-based phenotypic DST as the reference standard
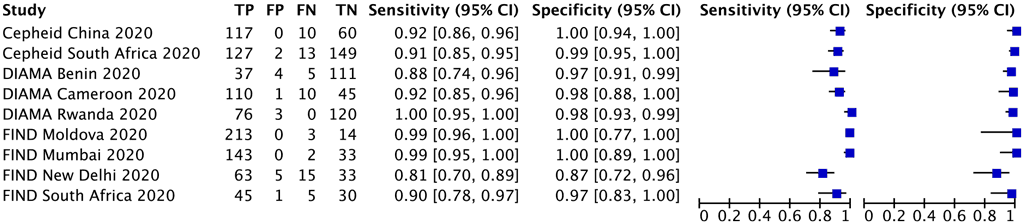
CI: confidence interval; DIAMA: Diagnostics for Multidrug Resistant Tuberculosis in Africa; DST: drug susceptibility testing; FIND: Foundation for Innovative New Diagnostics; FN: false negative; FP: false positive; TB: tuberculosis; TN: true negative; TP: true positive.
The sensitivity in these three studies ranged from 81% to 100% and the specificity from 87% to 100%. The pooled sensitivity was 94.2% (95% CI: 89.3–97.0%) and the pooled specificity was 98.0% (95% CI: 95.2–99.2%).
PICO 2: Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, irrespective of resistance to rifampicin, for detection of resistance to fluoroquinolones, as compared with culture-based phenotypic DST?
Three multinational studies with 1337 participants provided data for evaluation of detection of fluoroquinolone resistance. The reference standard for each of these studies was culturebased phenotypic DST. Each study centre in the multinational studies was analysed as a separate study (Fig. 2.4.1.2).
Specificity estimates were inconsistent, at 84% (FIND, Mumbai), 91% (FIND, New Delhi) and more than 96% for other studies. The heterogeneity in specificity estimates could not be explained. Consequently, the certainty of the evidence was downgraded one level for inconsistency; the quality (certainty) of the evidence was rated high for sensitivity and moderate for specificity.
Fig. 2.4.1.2 Forest plot of included studies for fluoroquinolone resistance detection, irrespective of rifampicin resistance with culture-based phenotypic DST as the reference standard
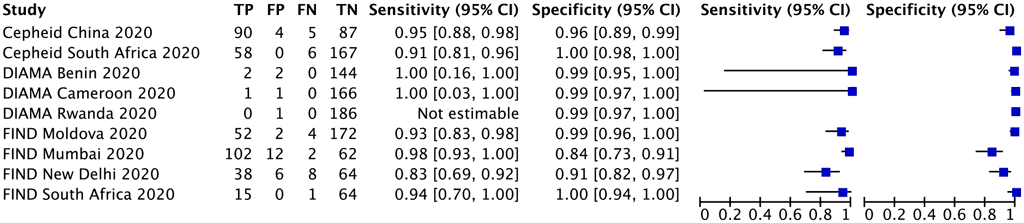
CI: confidence interval; DIAMA: Diagnostics for Multidrug Resistant Tuberculosis in Africa; DST: drug susceptibility testing; FIND: Foundation for Innovative New Diagnostics; FN: false negative; FP: false positive; TB: tuberculosis; TN: true negative; TP: true positive.
The sensitivity for fluoroquinolone resistance in these three studies ranged from 83% to 100% and the specificity from 84% to 100%. The pooled sensitivity was 93.1% (95% CI: 88.0–96.1%) and the pooled specificity was 98.3% (95% CI: 94.5–99.5%).
PICO 3: Should low complexity automated NAATs be used on culture isolates in people with signs and symptoms of pulmonary TB, and detected resistance to rifampicin, for detection of resistance to ethionamide, as compared with genotypic sequencing of the inhA promoter?
One multinational study with 434 participants provided data for evaluating resistance to ethionamide. The reference standard for this study was DNA sequencing of the inhA promoter. Each study centre in the multinational study was analysed as a separate study (Fig. 2.4.1.3).
The study was judged to be at very serious risk of bias in the reference standard domain because it did not include all loci (i.e. ethA, ethR and inhA promoter) required for the reference standard to classify the target condition correctly. Against a reference standard of phenotypic DST, the pooled sensitivity was considerably lower, at 51.7% (95% CI: 33.1–69.8%). Consequently, certainty of evidence was downgraded two levels for risk of bias for both sensitivity and specificity. In addition, the 95% CIs were wide for both sensitivity and specificity, which could lead to different decisions, depending on which confidence limits are assumed. Consequently, the certainty of the evidence was downgraded one level for imprecision for both sensitivity and specificity; the quality (certainty) of evidence was rated very low for both sensitivity and specificity.
Fig. 2.4.1.3 Forest plot of included studies for ethionamide resistance detection with genotypic DST as the reference standard
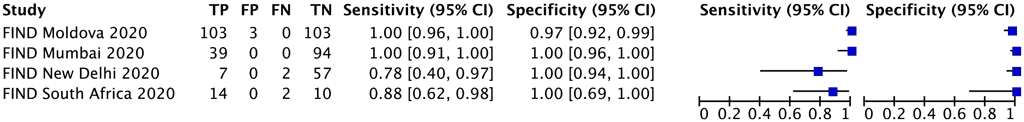
CI: confidence interval; DST: drug susceptibility testing; FIND: Foundation for Innovative New Diagnostics; FN: false negative; FP: false positive; TB: tuberculosis; TN: true negative; TP: true positive.
The sensitivity for ethionamide resistance in this study ranged from 78% to 100% and the specificity from 97% to 100%. The pooled sensitivity was 98.0% (95% CI: 74.2–99.9%) and the pooled specificity was 99.7% (95% CI: 83.5–100.0%).
PICO 4: Should low complexity automated NAATs be used on sputum in people with signs and symptoms of pulmonary TB, and detected resistance to rifampicin, for detection of resistance to amikacin, as compared with culture-based phenotypic DST?
One multinational study with 490 participants provided data for evaluating resistance to amikacin. The reference standard for this study was culture-based phenotypic DST. Each study centre in this multinational study was analysed as a separate study (Fig. 2.4.1.4).
The 95% CI for sensitivity was wide, which could lead to different decisions around true positives and false negatives, depending on which confidence limits are assumed. Also, there were few participants with amikacin resistance contributing to this analysis for the observed sensitivity. Consequently, the certainty of the evidence was downgraded two levels for imprecision. Also, there were few participants with amikacin resistance contributing to this analysis for the observed sensitivity. Consequently, the certainty of the evidence was downgraded two levels for imprecision; the quality (certainty) of evidence was rated low for sensitivity and high for specificity.
Fig. 2.4.1.4 Forest plot of included studies for amikacin resistance detection with culture-based phenotypic DST as the reference standard
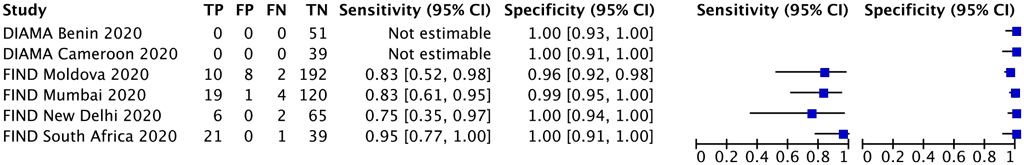
CI: confidence interval; DST: drug susceptibility testing; FIND: Foundation for Innovative New Diagnostics; FN: false negative; FP: false positive; TB: tuberculosis; TN: true negative; TP: true positive.
The sensitivity for amikacin resistance in this study ranged from 75% to 95% and the specificity from 96% to 100%. The pooled sensitivity was 86.1% (95% CI: 75.0–92.7%) and the pooled specificity was 98.9% (95% CI: 93.0–99.8%).
Cost–effectiveness analysis
This section answers the following additional question:
What is the comparative cost, affordability and cost–effectiveness of implementation of low complexity automated NAATs?
A systematic review was conducted, focusing on economic evaluations of low complexity automated NAATs. Four online databases (Embase, Medline, Web of Science and Scopus) were searched for new studies published from 1 January 2010 through 17 September 2020. The citations of all eligible articles, guidelines and reviews were reviewed for additional studies. Experts and test manufacturers were also contacted to identify any additional unpublished studies.
The objective of the review was to summarize current economic evidence and further understand the costs, cost–effectiveness and affordability of low complexity automated NAATs.
Two low complexity automated NAATs were identified: the MeltPro MTB/RIF (Xiamen Zeesan Biotech Co Ltd, China) and the Xpert MTB/XDR assay (Cepheid, Sunnyvale, USA). Only data concerning Xpert MTB/XDR are included in this review. As is the case with Xpert MTB/RIF, the novel XDR assay can be used to test either unprocessed or concentrated sputum. No published studies providing direct evidence on the cost or cost–effectiveness of low complexity automated NAATs were identified.
Through direct communication from the Xpert MTB/XDR manufacturer, Cepheid, the low- and middle-income country (LMIC) cost for the XDR cartridge is expected to be US$ 19.80 ex-works. Shipping and customs costs will be additional and will be borne by the ordering nations or organizations, as is currently the case for Xpert MTB/RIF and Ultra cartridges.
As with the Xpert MTB/RIF and Ultra assays, the test cartridge costs represent just one component of the total unit test costs that must be considered, with equipment being another important consideration. The Xpert MTB/XDR test will not work on existing six-colour modules and will require laboratories to upgrade to 10-colour GeneXpert modules. There will be different upgrade options for the 10-colour system, with different price points depending on the needs and resources available. Upgrade options include:
- a new 10-colour system – this is the most costly option, at US$ 9420 for one module to US$ 72 350 for 16 modules, including the GeneXpert platform, computer and scanner;
- a new 10-colour satellite instrument with the GeneXpert connected to an existing system – this costs from US$ 6495 for one module to US$ 69 525 for 16 modules; and
- converting an existing GeneXpert system from a six-colour to a 10-colour system by replacing modules – a 10-colour module kit costs US$ 3860.
Additional cost considerations for Xpert MTB/XDR include additional testing or repeated testing in the case of indeterminate or non-actionable results (indeterminate, non-determinate or invalid). The potential cost burden of this is likely to vary, depending on the proportion of indeterminate test results across settings and the associated re-testing protocols.
No studies that have directly assessed the cost–effectiveness of the Xpert MTB/XDR cartridge were identified. Although extrapolation from other platforms and testing approaches for costing may be appropriate, extrapolation of cost–effectiveness data from Xpert MTB/RIF (Ultra) or other NAATs is not advised because of differences in diagnostic accuracy, costs associated with XDR treatment, and the different testing and treatment cascade of care.
Several factors are likely to influence the cost–effectiveness of Xpert MTB/XDR; they include diagnostic accuracy, which may lead to more or fewer individuals being diagnosed compared with the standard of care (which in turn will vary, depending on the local standard of care). In addition to diagnostic accuracy associated with the test itself, the diagnostic algorithm and placement of the Xpert MTB/XDR test within the algorithm has important implications.
The novel Xpert MTB/XDR provides results in less than 90 minutes. Thus, introduction of this test is likely to result in faster time to a result for genotypic DST and could affect cost–effectiveness by improving the numbers of patients initiating treatment, reducing loss to follow-up and improving survival rates. Costs associated with XDR treatment are likely to be an important driver of cost and cost–effectiveness because previous work has shown that these costs are high compared to diagnostic and other treatment costs. As larger numbers of XDR-positive individuals requiring treatment are identified, total resources required to treat these individuals will increase.
In the absence of transmission modelling studies, there is no information on the long-term population level impact of introducing Xpert MTB/XDR. Nevertheless, the benefits of identifying more cases earlier could lead to a reduction in ongoing transmission and potential cost-savings over the long term. This requires thorough investigations through transmission modelling.
How large are the resource requirements (costs)?
No published studies provided direct evidence about the total resources required. Resource requirements will include the purchase of cartridges (US$ 19.80/cartridge), upgrading of existing platforms to 10-colour modules (an upgrade that will eventually be required for all Xpert platforms: US$ 3860 to >US$ 72 350) and operational and programmatic costs associated with implementing the novel diagnostic. Resource requirements for XDR treatment (e.g. drugs, hospital capacity and staff) are also likely to increase as the number of people diagnosed increases. Total costs will vary, depending on testing volume and prevalence of XDR in the population; also, the impact on the budget will depend on the current standard of care and associated resource use.
What is the certainty of the evidence of resource requirements (costs)?
Direct costs related to the purchase of cartridges and machinery are provided from the manufacturer; however, several important items related to resource use for implementing Xpert MTB/XDR have not been investigated (e.g. staff time, overhead and operational costs). Differences in resource use between Xpert MTB/XDR and existing approaches will vary across settings using different phenotypic and genotypic DST. There is important variability in costs of staff time and operational costs (e.g. testing volume) across settings.
Does the cost–effectiveness of the intervention favour the intervention or the comparison?
No cost–effectiveness studies using Xpert MTB/XDR were identified. Extrapolation of cost– effectiveness data from Xpert MTB/RIF or other NAATs is not advised because of differences in diagnostic accuracy, and costs associated with XDR treatment and the testing and treatment cascade of care.
More details on economic evidence synthesis and analysis are provided in Web Annex B.20: Systematic literature review of economic evidence for NAATs to detect TB and DR-TB in adults and children.
User perspective
This section answers the following question about key informants’ views and perspectives on the use of low complexity automated NAATs:
- Is there important uncertainty about or variability in how much end-users value the main outcomes?
- What would be the impact on health equity?
- Is the intervention acceptable to key stakeholders?
- Is the intervention feasible to implement?
The synthesis and analysis of qualitative evidence on end-users’ perspectives are discussed above in the section “User perspective” for moderate complexity automated NAATs: chapter 2.1.2 of the current guidelines.
Findings of the review and interviews
The main findings of the systematic review and interviews are given below. Where information is from the review, a level of confidence in the QES is given; where it is from interviews, this is indicated with ‘Interviews’.
Is there important uncertainty about or variability in how much end-users value the main outcomes?
- Patients in high burden TB settings value:
- getting an accurate diagnosis and reaching diagnostic closure (finally knowing “what is wrong with me”);
- avoiding diagnostic delays because they exacerbate existing financial hardships and emotional and physical suffering, and make patients feel guilty for infecting others (especially children);
- having accessible facilities; and
- reducing diagnosis-associated costs (e.g. travel, missing work) as important outcomes of the diagnostic.
QES: moderate confidence
- Low complexity automated NAATs, when compared with existing tests or sputum microscopy, are appreciated by health care professionals because of:
- the rapidity and accuracy of the results;
- the confidence that a result generates to start treatment and motivate patients;
- the diversity of sample types;
- the ability to detect drug resistance earlier or at all, for as many drugs as possible (altering a clinician’s risk perception of drug resistance in children), and the consequence of avoiding costlier investigations or hospital stays.
QES: high confidence - Compared with other available diagnostic methods, the cartridge has a quicker turnaround time for first- and second-line DST. Health care professionals value the faster turnaround time, the potential ability to reflex samples from the Xpert MTB/RIF to the Xpert MTB/XDR cartridge, and receiving information on multiple drugs and high-level or low-level resistance simultaneously, because it could enable quicker diagnosis and optimized treatment for patients.
Interviews
- Laboratory technicians appreciate low complexity automated NAATs for the following reasons:
- Overall, the tests improve laboratory work compared with sputum microscopy in terms of ease of use, ergonomics and biosafety.
QES: high confidence - These tests require minimal user steps, and the GeneXpert platform is a familiar system that people feel comfortable running and interpreting.
Interviews
- Overall, the tests improve laboratory work compared with sputum microscopy in terms of ease of use, ergonomics and biosafety.
- Laboratory managers appreciate that monitoring of laboratory work and training is easier than with sputum microscopy, and that use of low complexity automated NAATs eases staff retention because it increases staff satisfaction and is symbolic of progress within the TB world.
QES: low confidence
What would be the impact on health equity?
The impact on health equity would be similar to that of moderate complexity automated NAATs: Chapter 2.1.2 of the current guidelines.
Is the intervention acceptable to key stakeholders?
The acceptability to key stakeholders is similar to that of moderate complexity automated NAATs: Chapter 2.1.2 of the current guidelines.
The identified challenges in implementing the use of low complexity automated NAATs and accumulated delays at every step may compromise the added value and benefits identified by the users (e.g. avoiding delays, keeping costs low, accurate results, information on drug resistance and easing laboratory work), ultimately leading to use. QES: high confidence If these values are not met, it can be assumed that users are less likely to find low complexity automated NAATs acceptable.
Is the intervention feasible to implement?
- Low complexity automated NAATs may decrease the workload in the laboratory in terms of freeing up time for laboratory staff. However, based on experience with Xpert MTB/RIF (Ultra), the introduction of a new class of technologies may increase the workload of laboratory staff if added onto existing work without adjusting staffing arrangements or if the new technology does not replace existing diagnostic tests.
QES: moderate confidence - Low complexity automated NAATs require less user training than other DST methods (e.g. LPA and culture), making these tests more feasible to implement than methods with more user steps and those that require significant additional training.
Interview study
Implementation of new diagnostics must be accompanied by training for clinicians, to help them interpret results from new molecular tests and understand how this relates to the treatment of a patient. In the past, with the introduction of Xpert MTB/RIF (Ultra), this has been a challenge and has led to underuse.
QES: high confidence and interview study
Introduction of Xpert MTB/RIF (Ultra) has also led to overreliance on results of cartridge-based NAATs at the expense of clinical acumen.
QES: moderate confidence - Introduction of new diagnostics must also be accompanied by guidelines and algorithms that support clinicians and laboratories in communicating with each other; for example, these resources allow clinicians and laboratories to discuss discordant results, and interpret laboratory results in the context of drug availability, patient history and patient progress on a current drug regimen.
Interviews - An efficient sample transportation system, with sustainable funding mechanisms, is crucial for feasibility, especially if an algorithm requires multiple samples at different times from different collection points, as is the case when dealing with DR-TB. If mishandled during preparation, there is a risk that the sample may become contaminated and yield inconclusive results on molecular diagnostics. Participants cited good personnel skills, standardized operating procedures and significant laboratory infrastructure as essential in reducing sample contamination in their laboratory.
Interviews - The feasibility of low complexity automated NAATs is challenged if there is an accumulation of diagnostic delays or underuse (or both) at every step in the process, mainly because of health system factors:
- non-adherence to testing algorithms, testing for TB or MDR-TB late in the process, empirical treatment, false negatives due to technology failure, large sample volumes and staff shortages, poor or delayed sample transport and sample quality, poor or delayed communication of results, delays in scheduling follow-up visits and recalling patients, and inconsistent recording of results;
- lack of sufficient resources and maintenance (e.g. stock-outs; unreliable logistics; lack of funding, electricity, space, air conditioners and sputum containers; dusty environment; and delayed or absent local repair option);
- inefficient or unclear workflows and patient flows (e.g. inefficient organizational processes, poor links between providers, and unclear follow-up mechanisms or information on where patients need to go); and
- lack of data-driven and inclusive national implementation processes.
QES: high confidence
- The feasibility of using low complexity automated NAATs is also challenged by the value of diagnosing MTB over DR-TB at primary care. This situation makes the NAAT less feasible as a baseline test, although it would fit at a district or intermediate level laboratory.
Implementation considerations
Factors to consider when implementing low complexity automated NAATs for detection of resistance to isoniazid and second-line anti-TB agents are as follows:
- local epidemiological data on resistance prevalence should guide local testing algorithms, whereas pretest probability is important for the clinical interpretation of test results;
- the cost of a test varies depending on parameters such as the number of samples in a batch and the staff time required; therefore, a local costing exercise should be performed;
- low, moderate and high complexity tests have successive increase in technical competency needs (qualifications and skills) and staff time, which affects planning and budgeting;
- availability and timeliness of local support services and maintenance should be considered when selecting a provider;
- laboratory accreditation and compliance with a robust quality management system (including appropriate quality control) are essential for sustained service excellence and trust;
- training of both laboratory and clinical staff will ensure effective delivery of services and clinical impact;
- use of connectivity solutions for communication of results is encouraged, to improve efficiency of service delivery and time to treatment initiation;
- rapid and early testing for the detection of fluoroquinolone resistance is essential before starting treatment with the all-oral MDR/RR-TB shorter regimen (i.e. 6–9 months); this may also become relevant (depending on the epidemiological context) if new shorter drugsusceptible TB regimens that include fluoroquinolones are introduced;
- these tests can be used to rule in ethionamide resistance, but not to rule out resistance, because mutations conferring resistance to ethionamide are not limited to the inhA promoter region – they also include ethA, ethR and other genes;
- culture-based phenotypic DST may still be required, particularly among those with a high pretest probability of resistance when the low complexity automated NAATs does not detect drug resistance; in addition, culture-based phenotypic DST:
- remains important to determine resistance to other anti-TB agents, particularly the new and repurposed medicines, and to monitor the emergence of additional drug resistance;
- does not apply to ethionamide because it is unreliable and poorly reproducible;
- for second-line injectable drugs, the panel evaluated the performance in detecting resistance to amikacin only because both kanamycin and capreomycin are no longer recommended for the treatment of DR-TB; and
- culture-based phenotypic DST may be important to confirm amikacin susceptibility in situations where it is appropriate to use this medicine, to balance risk and benefit.
Research priorities
Research priorities for low complexity automated NAATs for detection of resistance to isoniazid and second-line anti-TB agents are as follows:
- diagnostic accuracy, in specific patient populations (e.g. children, people living with HIV, and patients with signs and symptoms of extrapulmonary TB) and in non-sputum samples;
- impact of diagnostic technologies on clinical decision-making and outcomes that are important to patients (e.g. cure, mortality, time to diagnosis and time to start treatment) in all patient populations;
- impact of specific mutations on treatment outcomes among people with DR-TB;
- use, integration and optimization of diagnostic technologies in the overall landscape of testing and care, as well as diagnostic pathways and algorithms;
- economic studies evaluating the costs, cost–effectiveness and cost–benefit of different diagnostic technologies;
- qualitative studies evaluating equity, acceptability, feasibility and end-user values of different diagnostic technologies;
- effect of non-actionable results (indeterminate, non-determinate or invalid) on diagnostic accuracy and outcomes that are important to patients;
- evaluation of low complexity automated NAATs for initial TB detection, in addition to its use as a follow-on test, in all people with signs and symptoms of TB, in children and in people living with HIV; and
- the potential utility of katG resistance detection to identify MDR-TB clones that may be missed because they do not have an RRDR mutation (e.g. the Eswatini MDR-TB clone, which has both the katG S315T and the non-RRDR rpoB I491F mutation).
2.4.2 First-line LPAs
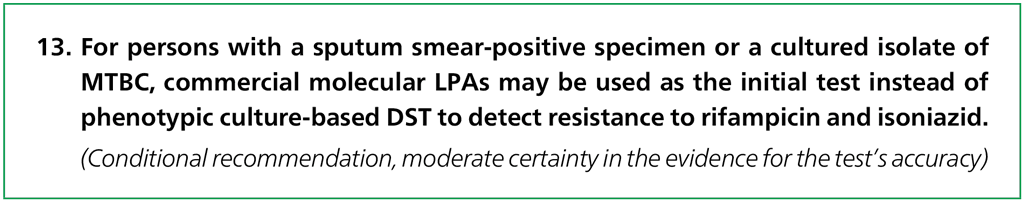
In 2008, WHO approved the use of commercial LPAs for detecting MTBC in combination with resistance to rifampicin and isoniazid in sputum smear-positive specimens (direct testing) and in cultured isolates of MTBC (indirect testing). A systematic review at that time evaluated the diagnostic accuracy of two commercially available LPAs – the INNO-LiPA Rif.TB assay (Innogenetics, Ghent, Belgium), and the GenoType® MTBDRplus (version 1), hereafter referred to as Hain version 1 – and provided evidence for WHO’s endorsement (37, 38). Excellent accuracy was reported for both tests in detecting rifampicin resistance, but their diagnostic accuracy for isoniazid resistance had lower sensitivity, despite the high specificity. Because there were inadequate data to allow stratification by smear status, WHO’s recommendation for using LPAs was limited to culture isolates or smear-positive sputum specimens. Further data have since been published on the use of LPAs; newer versions of LPA technology have now been developed, such as the Hain GenoType MTBDRplus version 2, hereafter referred to as Hain version 2; and other manufacturers have entered the market, including Nipro (Tokyo, Japan), which developed the Genoscholar™ NTM+MDRTB II, hereafter referred to as Nipro.
In 2015, FIND evaluated the Nipro and the Hain version 2 LPAs, and compared them with Hain version 1. The study demonstrated equivalence among the three commercially available LPAs for detecting TB and resistance to rifampicin and isoniazid (5).
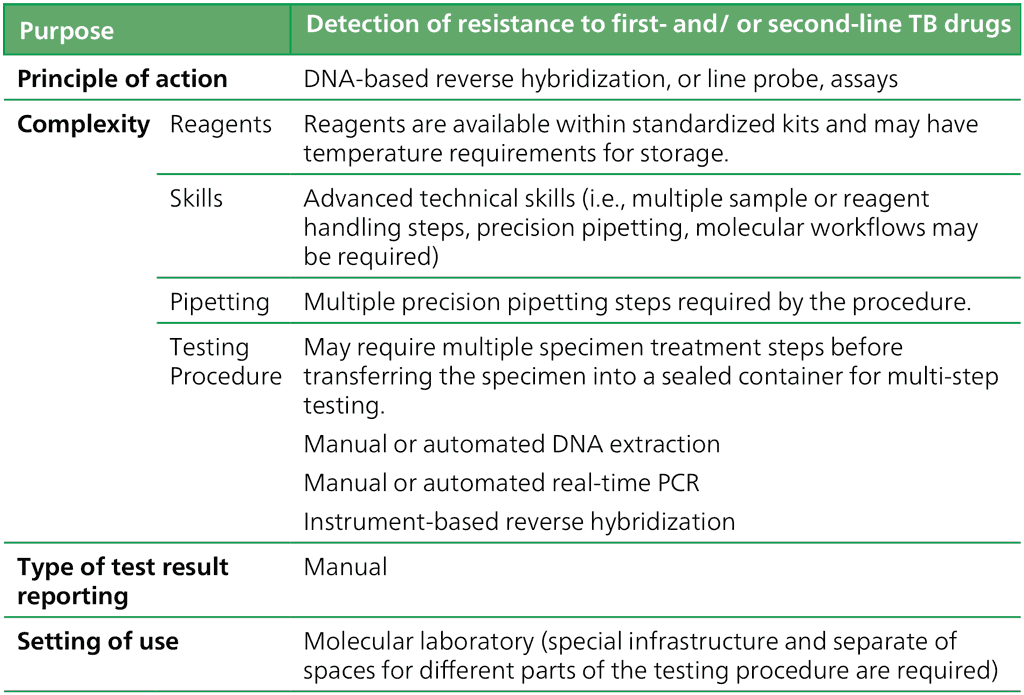
Table 2.4.2.1 Class criteria for LPAs
Recommendation
Remarks
- These recommendations apply to the use of LPAs for testing sputum smear-positive specimens (direct testing) and cultured isolates of MTBC (indirect testing) from both pulmonary and extrapulmonary sites.
- LPAs are not recommended for the direct testing of sputum smear-negative specimens.
- These recommendations apply to the detection of MTBC and the diagnosis of MDR-TB, but acknowledge that the accuracy of detecting resistance to rifampicin and isoniazid differs and, hence, that the accuracy of a diagnosis of MDR-TB is reduced overall.
- These recommendations do not eliminate the need for conventional culture-based DST, which will be necessary to determine resistance to other anti-TB agents and to monitor the emergence of additional drug resistance.
- Conventional culture-based DST for isoniazid may still be used to evaluate patients when the LPA result does not detect isoniazid resistance. This is particularly important for populations with a high pretest probability of resistance to isoniazid.
- These recommendations apply to the use of LPA in children based on the generalization of data from adults.
Test description
LPAs are a family of DNA strip-based tests that can detect the MTBC strain and determine its drug resistance profile through the pattern of binding of amplicons (DNA amplification products) to probes targeting the following: specific parts of the MTBC genome (for MTBC detection), the most common resistance-associated mutations to first-line and second-line agents, or the corresponding wild-type DNA sequence (for detection of resistance to anti-TB drugs) (38).
LPAs are based on reverse hybridization DNA strip technology and involve three steps: DNA extraction from M. tuberculosis culture isolates or directly from patient specimens, followed by multiplex PCR amplification and then reverse hybridization with visualization of amplicon binding (or lack thereof) to wild-type and mutation probes (8).
Although LPAs are more technically complex to perform than the Xpert MTB/RIF assay, they can detect isoniazid resistance. Testing platforms have been designed for a reference laboratory setting and are thus most applicable to high TB burden countries. Results can be obtained in 5 hours.
Some of these steps can be automated, making the method quicker and more robust, and reducing the risk of contamination.
The Hain version 1 and version 2 assays include rpoB probes to detect rifampicin resistance, katG probes to detect mutations associated with high-level isoniazid resistance, and inhA promoter probes to detect mutations usually associated with low-level isoniazid resistance. The probes used to detect wild-type and specific mutations are the same for both versions of the Hain LPA.
Similarly, the Nipro assay allows for the identification of MTBC, and resistance to rifampicin and isoniazid. The Nipro assay also differentiates M. avium, M. intracellulare and M. kansasii from other non-tuberculous mycobacteria.
The rpoB, katG and inhA promoter mutation probes are the same for the three assays, with the exception of the katG S315N mutation, which is included in the Nipro assay but not in Hain version 1 or version 2. There are some minor variations in the codon regions covered for the wild type among Hain version 1 and version 2, and the Nipro.
Justification and evidence
In 2015, WHO commissioned an updated systematic review of the accuracy of commercial LPAs for detecting MTBC, and resistance to rifampicin and isoniazid. A total of 74 studies were identified, comprising 94 unique datasets (see Web Annex A.7: “FL-LPA”). Of these 94 datasets, 83 evaluated Hain version 1, five evaluated Hain version 2, and six evaluated the Nipro assay. Only one of the studies performed head-to-head testing of all three target LPAs on directly tested clinical specimens and indirectly tested isolates, and these data were included as six separate datasets (9). No studies performed LPA testing on specimens and culture isolates from the same patients, precluding direct within-study comparisons.
Following the 2015 systematic review, the WHO Global TB Programme convened a GDG in March 2016 to assess the data and update the 2008 policy recommendations on using commercial LPAs to detect MTBC, and resistance to isoniazid and rifampicin. The PICO questions are given in Box 2.4.2.1.
LPAs were compared with a phenotypic culture-based DST reference standard, and a composite reference standard that combined the results from genetic sequencing with results from phenotypic culture-based DST. Phenotypic DST was the primary reference standard applied to all participants for all analyses. These analyses were stratified – first, by susceptibility or resistance to rifampicin or isoniazid (or both) and second, by type of LPA testing (indirect testing or direct testing).
Several studies contributed to either sensitivity (no true positives and no false negatives) or specificity (no true negatives and no false positives) but not to both. For these studies, a univariate, random-effects meta-analysis of the estimates of sensitivity or specificity was performed separately, to make optimal use of the data. The results from the univariate analysis (using all studies) were compared with the results from the bivariate analysis of the subset of studies that contributed to estimates of both sensitivity and specificity.
If there were at least four studies for index tests with data that contributed only to sensitivity or specificity, a univariate, random-effects meta-analysis was performed to assess one summary estimate, assuming no correlation between sensitivity and specificity. In cases in which there were fewer than four studies, or where substantial heterogeneity was evident on forest plots that precluded a meta-analysis, a descriptive analysis was performed for these index tests. Forest plots were visually assessed for heterogeneity among the studies within each index test and in the summary plots, for variability in estimates and the width of the prediction region (a wider prediction region suggests more heterogeneity).
Implementation considerations
Adopting LPAs to detect rifampicin and isoniazid resistance does not eliminate the need for conventional culture and DST capacity. Culture and phenotypic culture-based DST have critical roles in monitoring patients’ responses to treatment and detecting additional resistance to second-line agents.
- The adoption of LPA should be phased in, starting at national or central reference laboratories, or those with proven capability to conduct molecular testing. Expansion could be considered, within the context of a country’s plans for laboratory strengthening, the availability of suitable personnel in peripheral centres and the quality of specimen transport systems.
- Adequate and appropriate laboratory infrastructure and equipment should be provided, to ensure that the required precautions for biosafety and the prevention of contamination are met – specimen processing for culture and procedures for manipulating cultures must be performed in biological safety cabinets in TB-containment laboratories.
- Laboratory facilities for LPAs require at least three separate rooms, one each for DNA extraction, pre-amplification procedures, and amplification and post-amplification procedures. To avoid contamination, access to molecular facilities must be restricted, a unidirectional workflow must be implemented and stringent cleaning protocols must be established.
- Appropriate laboratory staff should be trained to conduct LPA procedures. Staff should be supervised by a senior staff member with adequate training and experience in molecular assays. A programme for the external quality assessment of laboratories using LPAs should be developed as a priority.
- Mechanisms for rapidly reporting LPA results to clinicians must be established, to provide patients with the benefit of early diagnosis. The same infrastructure used for performing LPAs can be used also to perform second-line LPAs.
- LPAs are designed to detect TB and resistance to rifampicin and isoniazid in the direct testing of processed sputum samples, and in the indirect testing of culture isolates of MTBC. The use of LPAs with other respiratory samples (e.g. from BAL or gastric aspiration) or extrapulmonary samples (e.g. tissue samples, CSF or other body fluids) have not been adequately evaluated.
- The availability of second-line agents is critical in the event that resistance to rifampicin or isoniazid, or both, is detected.
- For patients with confirmed MDR/RR-TB, second-line LPAs are recommended to detect additional resistance to second-line anti-TB agents.
Research priorities
- Development of improved understanding of the correlation between the detection of resistance-conferring mutations using culture-based DST and patient outcomes.
- Review of evidence to confirm or revise different critical concentrations used in culture-based DST methods.
- Determination of the limit of detection for LPA in detecting heteroresistance.
- Determination of needs for training, assessing competency and ensuring quality assurance.
- Gathering of more evidence on the impact on mortality of initiating appropriate treatment for MDR-TB.
- Meeting the STARD for future diagnostic studies.
- Performance of country-specific cost–effectiveness and cost–benefit analyses of LPA use in different programmatic settings.
2.4.3 Second-line LPAs
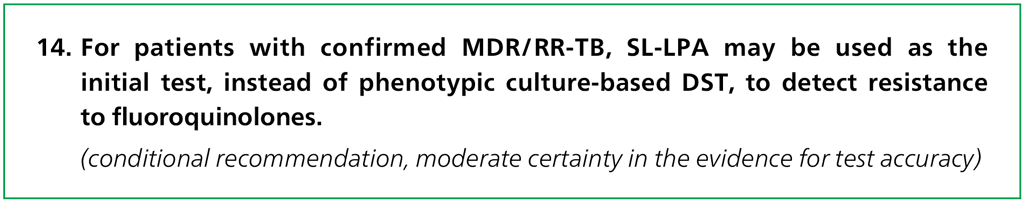
Genotypic (molecular) methods have considerable advantages for scaling up programmatic management and surveillance of DR-TB, offering rapid diagnosis, standardized testing, potential for high throughput and fewer requirements for laboratory biosafety. Molecular tests for detecting drug resistance – for example, the GenoType MTBDRsl assay (Hain Lifescience, Nehren, Germany), hereafter referred to as MTBDRsl (10) – have shown promise for the diagnosis of DR-TB. These tests are rapid (can be performed in a single working day) and detect the presence of mutations associated with drug resistance. MTBDRsl belongs to a category of molecular genetic tests called second-line LPAs (SL-LPAs).
MTBDRsl (version 1.0) was the first commercial SL-LPA for detection of resistance to secondline TB drugs. In 2015, the manufacturer developed and made commercially available version 2.0 of the MTBDRsl assay. Version 2.0 detects the mutations associated with fluoroquinolones and second-line injectable drug (SLID) resistance detected by version 1.0, and additional mutations. Once a diagnosis of MDR/RR-TB has been established, an SL-LPA can be used to detect additional resistance to second-line drugs.
The MTBDRsl assay incorporates probes to detect mutations within genes that are associated with resistance to either fluoroquinolones or SLIDs (gyrA and rrs for version 1.0 and those genes plus gyrB and the eis promoter for version 2.0). The presence of mutations in these regions does not necessarily imply resistance to all the drugs within a particular class. Although specific mutations within these regions may be associated with different levels of resistance (i.e. different minimum inhibitory concentrations) to each drug within these classes, the extent of cross-resistance is not completely understood.
Recommendations

Remarks
- These recommendations apply to the use of SL-LPA for testing sputum specimens (direct testing) and cultured isolates of M. tuberculosis (indirect testing) from both pulmonary and extrapulmonary sites. Direct testing on sputum specimens allows for the earlier initiation of appropriate treatment.
- These recommendations apply to the direct testing of sputum specimens from MDR/RR-TB, irrespective of the smear status, while acknowledging that the indeterminate rate is higher when testing smear-negative sputum specimens than with smear-positive sputum specimens.
- These recommendations do not eliminate the need for conventional phenotypic DST capacity, which will be necessary to confirm resistance to other drugs and to monitor the emergence of additional drug resistance.
- Conventional phenotypic DST can still be used in the evaluation of patients with negative SL-LPA results, particularly in populations with a high pretest probability for resistance to fluoroquinolones or SLID (or both).
- These recommendations apply to the use of SL-LPA in children with confirmed MDR/RR-TB, based on the generalization of data from adults.
- Resistance-conferring mutations detected by SL-LPA are highly correlated with phenotypic resistance to ofloxacin and levofloxacin.
- Resistance-conferring mutations detected by SL-LPA are highly correlated with phenotypic resistance to SLID.
- Given the high specificity for detecting resistance to fluoroquinolones and SLID, the positive results of SL-LPA could be used to guide the implementation of appropriate infection control precautions.
Test description
The SL-LPA is based on the same principle as the first-line LPA. The assay procedure can be performed directly using a processed sputum sample or indirectly using DNA isolated and amplified from a culture of M. tuberculosis. Direct testing involves the following steps:
- Decontamination (e.g. with sodium hydroxide) and concentration of a sputum specimen by centrifugation.
- Isolation and amplification of DNA.
- Detection of the amplification products by reverse hybridization.
- Visualization using a streptavidin-conjugated alkaline phosphatase colour reaction.
Indirect testing includes only Steps 2–4. The observed bands, each corresponding to a wild-type or resistance-genotype probe, can be used to determine the drug susceptibility profile of the analysed specimen. The assay can be performed and completed within a single working day. Further details on the test process and practical support for implementation can be found in the WHO operational handbook. Module 3: diagnosis.
The index test used was MTBDRsl versions 1.0 and 2.0. These SL-LPAs detect specific mutations associated with resistance to the class of fluoroquinolones (including ofloxacin, levofloxacin, moxifloxacin and gatifloxacin) and SLIDs (including kanamycin, amikacin and capreomycin) in the MTBC. The MTBDRsl LPA detects mutations in the gyrA quinolone resistance-determining region (codons 85–97) and rrs (codons 1401, 1402 and 1484), and version 2.0 of the test added detection of mutations in the gyrB quinolone resistance-determining region (codons 536–541) and the eis promoter region (codons –10 to –14) (40). Version 2.0 is therefore expected to have improved sensitivity for resistance detection to these classes of drugs. Lastly, while version 1.0 included detection of mutations in embB that may encode for resistance to ethambutol, it was omitted from version 2.0 due to its status as a first line anti-TB drug. Therefore, this review did not determine the accuracy for ethambutol resistance.
More data are needed to better understand the correlation of the presence of certain fluoroquinolone resistance-conferring mutations with phenotypic DST resistance and with patient outcomes.
Justification and evidence
In March 2016, the WHO Global TB Programme convened a GDG to assess available data on the use of the MTBDRsl assay. WHO commissioned a systematic review on the accuracy and clinical use of assays for the detection of mutations associated with resistance to fluoroquinolones and SLID in people with MDR/RR-TB.
The PICO questions in Box 2.4.3.1 were designed to form the basis for the evidence search, retrieval and analysis.
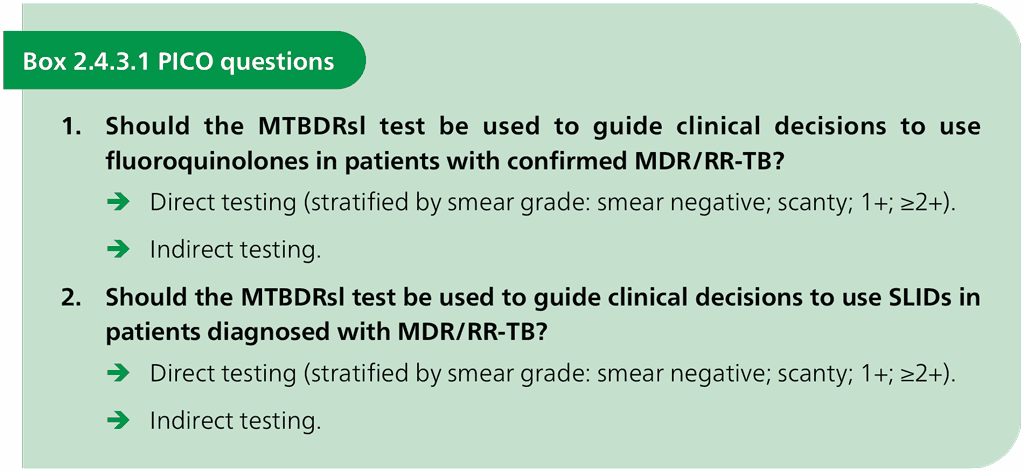
Twenty-nine unique studies were identified; of these, 26 evaluated the MTBDRsl version 1.0 assay (including 21 studies from the original Cochrane review). Three studies (one published and two unpublished) evaluated version 2.0. Data for version 1.0 and version 2.0 of the MTBDRsl assay were analysed separately. A phenotypic culture-based DST reference standard was used for the primary analyses. These analyses were stratified first by susceptibility or resistance to a particular drug, and second by type of SL-LPA testing (indirect testing or direct testing).
Performance of SL-LPA on sputum specimens and culture isolates
In patients with MDR/RR-TB, a positive SL-LPA result for fluoroquinolone resistance (as a class) or SLID resistance (as a group) can be treated with confidence. The diagnostic accuracy of SL-LPA is similar when performed directly on sputum specimens or indirectly on cultured isolates of M. tuberculosis.
Given the confidence in a positive result and the ability of the test to provide rapid results, the GDG felt that SL-LPA may be considered for use as an initial test for resistance to the fluoroquinolones and when relevant SLIDs. However, when the test shows a negative result, phenotypic culture-based DST may be necessary, especially in settings with a high pretest probability for resistance to either fluoroquinolones or SLIDs (or both). The use of SL-LPA in routine care should improve the time to the diagnosis of fluoroquinolone and where relevant SLIDs, especially when used for the direct testing of sputum specimens of patients with confirmed MDR/RR-TB. Early detection of drug resistance should allow for the earlier initiation of appropriate patient therapy and improved patient health outcomes. Overall, the test performs well in the direct testing of sputum specimens from patients with confirmed MDR/RR-TB, although the indeterminate rate is higher when testing smear-negative sputum specimens compared with smear-positive sputum specimens.
When the MTBDRsl assay is used in the direct testing of smear-negative sputum specimens from a population of patients with confirmed DR-TB, up to 44% of the results may be indeterminate (less with version 2.0, although very limited data) and hence require repeat or additional testing. However, if the same test were to be applied to the testing of smear-negative sputum specimens from patients without confirmed TB or DR-TB (i.e. patients suspected of having DR-TB), the indeterminate rate for the test would be significantly higher. Given the test’s sensitivity and specificity when an SL-LPA is done directly on sputum, the GDG felt that SL-LPAs can be used for the testing of all sputum specimens from patients with confirmed MDR/RR-TB, irrespective of whether the microscopy result is positive or negative.
For the reasons mentioned above (inadequate data owing to too few studies on version 2.0), results are not presented here for version 2.0. For MTBDRsl version 2.0, the data were either too sparse or too heterogeneous to combine in a meta-analysis or to compare indirect and direct testing.
Three studies evaluated the MTBDRsl version 2.0 in 562 individuals, including 111 confirmed cases of TB with fluoroquinolone resistance by indirect testing on a culture of M. tuberculosis compared with a phenotypic culture-based DST reference standard. Estimates of sensitivity ranged from 84% to 100% and specificity from 99% to 100%.
See Web Annex B.15: Drug concentrations used in culture-based DST SL-LPA for details of the drug concentrations used in culture-based DST to evaluate the performance of SL-LPAs in each included study.
Implementation considerations
The SL-LPA should only be used to test specimens from patients with confirmed MDR/RR-TB. Adoption of SL-LPAs does not eliminate the need for conventional culture and DST capability. Despite good specificity of SL-LPAs for the detection of resistance to fluoroquinolones and the SLIDs, culture and phenotypic DST is required to completely exclude resistance to these drug classes as well as to other second-line drugs. The following implementation considerations apply:
- SL-LPAs cannot determine resistance to individual drugs in the class of fluoroquinolones. Resistance-conferring mutations detected by SL-LPAs are highly correlated with phenotypic resistance to ofloxacin and levofloxacin.
- Mutations in some regions (e.g. the eis promoter region) may be responsible for causing resistance to one drug in a class more than other drugs within that class. For example, the eis C14T mutation is associated with kanamycin resistance in strains from Eastern Europe.
- SL-LPAs should be used in the direct testing of sputum specimens, irrespective of whether samples are smear negative or smear positive.
- SL-LPAs are designed to detect TB and resistance to fluroquinolones and SLIDs from sputum samples. Other respiratory samples (e.g. BAL and gastric aspirates) or extrapulmonary samples (tissue samples, CSF or other body fluids) have not been adequately evaluated.
- Culture and phenotypic DST plays a critical role in the monitoring of a patient’s response to treatment, and in detecting additional resistance to second-line drugs during treatment.
- SL-LPAs are suitable for use at the central or national reference laboratory level; they can also be used at the regional level if the appropriate infrastructure can be ensured (three separate rooms are required).
- All patients identified by SL-LPAs should have access to appropriate treatment and ancillary medications.
Research priorities
- Development of improved understanding of the correlation between the detection of resistance-conferring mutations with phenotypic DST results and with patient outcomes.
- Development of improved knowledge of the presence of specific mutations detected with SL-LPA correlated with minimum inhibitory concentrations for individual drugs within the classes of fluoroquinolones and SLIDs.
- Determination of the limit of detection of SL-LPA for the detection of heteroresistance.
- Gathering of more evidence on the impact of MTBDRsl on appropriate MDR-TB treatment initiation and mortality.
- Strongly encourage that future studies follow the recommendations in the STARD (11) statement to improve the quality of reporting.
- Performance of country-specific cost–effectiveness and cost–benefit analyses of the use of SL-LPA in different programmatic settings.
2.4.4 High complexity reverse hybridization-based NAATs for detection of pyrazinamide resistance
Pyrazinamide is an important antibiotic for the treatment of both drug-susceptible TB and DR-TB because of its unique ability to eradicate persisting bacilli and its synergistic properties with other antibiotics. Mono-resistance to pyrazinamide is rare; however, pyrazinamide resistance is strongly associated with MDR/RR-TB, with an estimated 30–60% of MDR/RR-TB also resistant to pyrazinamide. Thus, for people diagnosed with RR-TB, it is important to detect the presence of pyrazinamide resistance so that clinicians can make an informed decision on whether to include or exclude pyrazinamide in the treatment regimen. The high complexity hybridization-based NAAT may be used for diagnosis of pyrazinamide resistance on patient isolates; however, performance of this test requires appropriate infrastructure and skilled staff.
Recommendation

In terms of subgroups to be considered for this recommendation, no special considerations are required (e.g. for children, people living with HIV and those with extrapulmonary TB), given that the test is recommended for use on culture isolates.
Test description
Nipro (Osaka, Japan) developed Genoscholar™ PZA-TB, an LPA with reverse hybridization-based technology for detection of pyrazinamide resistance (12). This assay is a commercially available rapid molecular test for detection of pyrazinamide resistance. Compared with MTBDRplus and MTBDRsl LPA, the Genoscholar PZA-TB LPA does not include specific mutant probes because resistance mutations are widespread across the entire pncA gene with no predominant mutations. Instead, the Genoscholar PZA-TB assay targets a 700 base pair (bp) fragment covering the entire pncA gene and promoter region up to nucleotide –18 of the wild-type H37Rv reference strain.
Fig. 2.4.4.1 Nipro GenoScholar PZA-TB II strip (a) and Nipro GenoScholar PZA-TB II kit contents (b)
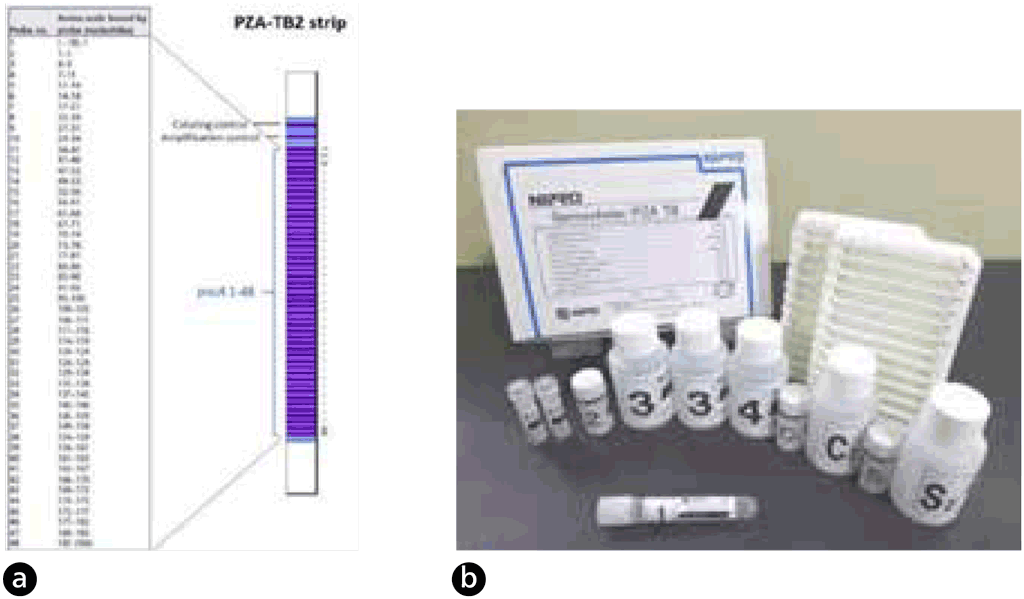
DNA extracted from cultures is amplified with primers by PCR. Amplified DNA is then hybridized to complementary oligonucleotide probes that are bound on a membrane strip. Streptavidin labelled with alkaline phosphatase is then added, to bind to any hybrids formed in the previous step. Next, a substrate is added, and an enzymatic reaction results in purple bands, which are visually interpreted. The absence of wild-type probe binding indicates the presence of a mutation. The first version of the assay contained 47 probes, which covered the pncA promoter and open reading frame. The second version contained 48 probes, three of which (pncA 16, 17 and 35) represent silent mutations known to be genetic markers not associated with pyrazinamide resistance: Gly60Gly (probe 16), Ser65Ser (probe 17) and Thr142Thr (probe 35).
Justification and evidence
The Genoscholar PZA-TB LPA assay, which is already commercially available, could potentially be implemented for diagnosis of pyrazinamide resistance in routine care. However, limited data have been published on the diagnostic accuracy of the assay. This systematic review with meta-analysis aimed to assist in collating all the available data to understand the diagnostic accuracy of the pyrazinamide LPA assay for detection of pyrazinamide resistance in TB patients, to guide policy-makers and clinicians.
The WHO Global TB Programme initiated an update of the current guidelines and commissioned a systematic review on the use of high complexity reverse hybridization-based NAATs for detection of pyrazinamide resistance in people with signs and symptoms of TB.
Two PICO questions were designed to form the basis for the evidence search, retrieval and analysis:
- Should high complexity reverse hybridization-based NAATs on sputum be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST or composite reference standard?
- Should high complexity reverse hybridization-based NAATs on isolates be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST?
The databases searched were PubMed, Web of Science and Embase, and they were searched without language or date restrictions. The search query was (PZA OR pyrazinamide OR pncA) AND (tuberculosis) AND (“line-probe assay” OR LPA OR “hybridization-based technology”). In addition, we approached Nipro (Osaka, Japan) to identify non-published data.
The microbiological reference standard was defined either as phenotypic culture-based DST performed using BD MGIT 960 PZA liquid assay or another acceptable phenotypic assay, or as genotypic DST performed using either targeted sequencing of the pncA gene or whole genome sequencing. In the case of genotypic DST, all samples with a pncA wild type were defined as being susceptible, while any variant in pncA was considered resistant, which implicitly would categorize “silent” mutations as resistant. In contrast, the composite reference standard was defined by classifying all samples with pncA wild type, pncA silent mutations and neutral mutations as being susceptible, while any other variant in pncA was considered resistant (13).
Data synthesis was structured around the two preset PICO questions, as outlined below. Three web annexes give additional information, as follows:
- details of studies included in the current analysis (Web Annex A.9: High complexity reverse hybridization-based NAATs);
- a summary of the results and details of the evidence quality assessment (Web Annex A.9: High complexity reverse hybridization-based NAATs); and
- a summary of the GDG panel judgements (Web Annex A.9: High complexity reverse hybridization-based NAATs).
PICO 1: Should high complexity reverse hybridization-based NAATs on sputum be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST or composite reference standard?
Three studies with a total of 122 participants provided data for evaluation of these NAATs for detection of pyrazinamide resistance, including two studies (101 participants) with phenotypic culture-based reference standard and one study (21 participants) with genotypic reference standard. The number of studies and participants were considered insufficient to make a conclusion on a diagnostic accuracy of high complexity reverse hybridization-based NAATs on sputum.
PICO 2: Should high complexity reverse hybridization-based NAATs on isolates be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST?
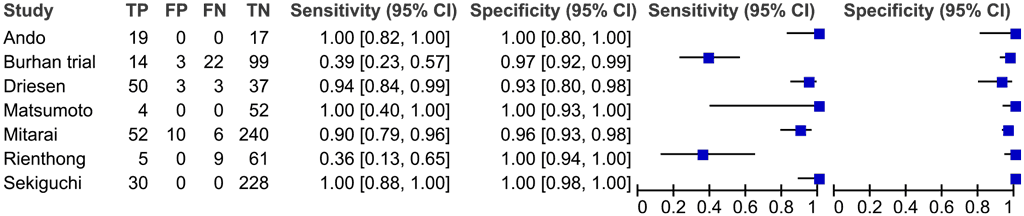
Seven studies with a total of 964 participants provided data for evaluation of these NAATs for detection of pyrazinamide resistance compared with a phenotypic culture-based reference standard (Fig. 2.4.4.2).
The studies suffered from selection bias because they selected isolates with a wide range of different pncA mutations rather than a representative sample from a population. Thus, the evidence was downgraded by one level for risk of bias. The included studies did not directly address the review question; hence, the evidence was downgraded one level for indirectness. The Burhan trial and the Rienthong study are outliers for their sensitivities compared with the other studies; hence, the evidence was downgraded one level for inconsistency. Taking these judgements together, the quality (certainty) of evidence was rated very low for sensitivity and low for specificity.
Fig. 2.4.4.2 Forest plot of included studies for pyrazinamide resistance detection, irrespective of rifampicin resistance with culture-based phenotypic DST as the reference standard
CI: confidence interval; DST: drug susceptibility testing; FN: false negative; FP: false positive; TB: tuberculosis; TN: true negative; TP: true positive.
The overall sensitivity for pyrazinamide resistance in these seven studies ranged from 36% to 100% and the specificity from 96% to 100%. The pooled sensitivity was 81.2% (95% CI: 75.4–85.8%) and specificity was 97.8% (95% CI: 96.5–98.6%).
More details on diagnostic accuracy of the high complexity reverse hybridization-based NAATs, including comparison with genotypic and composite reference standards are available in Web Annex 4.17: High complexity reverse hybridization-based NAATs: diagnostic accuracy for detection of resistance to pyrazinamide. A systematic review.
Cost–effectiveness analysis
This section answers the following additional question:
What is the comparative cost, affordability and cost–effectiveness of implementation of high complexity reverse hybridization-based NAATs?
A systematic review was carried out, focusing on economic evaluations of high complexity reverse hybridization-based NAATs. Four online databases (Embase, Medline, Web of Science and Scopus) were searched for new studies published from 1 January 2010 through 17 September 2020. The citations of all eligible articles, guidelines and reviews were reviewed for additional studies. The experts and test manufacturers were also contacted to identify any additional unpublished studies.
The objective of the review was to summarize current economic evidence and further understand the costs, cost–effectiveness and affordability of high complexity reverse hybridization-based NAATs.
No published studies were identified assessing costs or cost–effectiveness using the commercially available high complexity hybridization-based NAAT (Genoscholar PZA-TB II, Nipro Japan). Indirect evidence was available from several sources. Four studies examining other commercially available LPAs (Genotype MTBDRsl and MTBDRplus, Hain Lifescience) were identified.
The Genoscholar PZA LPA was developed for use with the Nipro automated MultiBlot; however, a recent unpublished trial¹² demonstrated that the Twincubator by Hain Lifescience could be used successfully with this LPA. This finding could make it easier to implement the Genoscholar PZA LPA in selected settings where Hain Lifescience equipment is already in use.
How large are the resource requirements (costs)?
No direct evidence from published studies was found regarding the total resources required. Resource requirements will include the purchase of test kits (Genoscholar PZA LPA: US$ 16/test kit consumables only), and the equipment, which is available for US$ 14 000. Operational costs are frequently several times greater than test kit costs (and will vary across settings), but are not accounted for usually. Nipro hopes that further reductions in test costs can be achieved when the Genoscholar PZA-TB II product is distributed globally.
Unit test costs for the Genotype MTBDRsl and MTBDRplus ranged from US$ 23.46 to US$ 108.70 (14–15), with higher unit test costs in countries such as China and South Africa, largely driven by higher staff wages and operational costs. Extrapolations from unit test costs using different LPAs should be done with caution, and they are not intended to be directly transferrable estimates. Nevertheless, these indirect data do suggest that the total unit test cost of the Genoscholar PZA-TB II is likely several-fold higher than the unit test kit consumable cost of US$ 16.
Total costs will vary, depending on testing volume, numbers eligible for testing and prevalence of pyrazinamide resistance in the population. The impact on the budget will depend on the current standard of care, diagnostic and care pathways, and associated resource use.
What is the certainty of the evidence of resource requirements (costs)?
Direct costs related to test kits and machinery are available, whereas several important items related to resource use (e.g. staff time, and overhead and operational costs associated with implementing Genoscholar PZA-TB II) have not been investigated. Differences in resource use between Genoscholar PZA-TB II and existing approaches will vary across settings that are using different phenotypic and genotypic DST. Also, there is important variability in costs of staff time and operation (e.g. testing volume) across settings.
Does the cost–effectiveness of the intervention favour the intervention or the comparison?
No cost–effectiveness studies were identified using the Genoscholar PZA-TB II. Extrapolation of cost–effectiveness data from other LPAs is not advised owing to differences in diagnostic accuracy, resistance prevalence, and the testing and treatment cascade of care.
More details on economic evidence synthesis and analysis are given in Web Annex 4.9: Systematic literature review of economic evidence for NAATs to detect TB and DR-TB in adults and children.
User perspective
This section answers the following questions about key informants’ views and perspectives on the use of high complexity reverse hybridization-based NAATs:
- Is there important uncertainty about or variability in how much end-users value the main outcomes?
- What would be the impact on health equity?
- Is the intervention acceptable to key stakeholders?
- Is the intervention feasible to implement?
Findings of the review and interviews
The main findings of the systematic review and interviews are given below. Where information is from the review, a level of confidence in the QES is given; where it is from interviews, this is indicated with ‘Interviews’.
Is there important uncertainty about or variability in how much end-users value the main outcomes?
- Patients in high burden TB settings value:
- getting an accurate diagnosis and reaching diagnostic closure (finally knowing “what is wrong with me”);
- avoiding diagnostic delays because they exacerbate existing financial hardships and emotional and physical suffering, and make patients feel guilty for infecting others (especially children);
- having accessible facilities; and
- reducing diagnosis-associated costs (e.g. travel, missing work) as important outcomes of the diagnostic.
QES: moderate confidence
- The high complexity reverse hybridization-based NAATs meet some preferences and values of laboratory staff and clinicians, in that the current test:
- provides quicker results about pyrazinamide resistance than other available methods (e.g. culture DST);
- can provide information on different concentration levels; and
- targets a drug that is widely used in first-line TB treatment.
Interviews
What would be the impact on health equity?
The impact on health equity would be similar to that of moderate complexity automated NAATs (Section 2.1.2), plus the following:
- Lengthy diagnostic delays, underuse of diagnostics, lack of TB diagnostic facilities at lower levels and too many eligibility restrictions hamper access to prompt and accurate testing and treatment, particularly for vulnerable groups.
QES: high confidence
Applicability to three index tests also confirmed in interviews - Staff and managers voiced concerns about the sustainability of funding and maintenance, complex conflicts of interest between donors and implementers, and the strategic and equitable use of resources, which makes it difficult to ensure equitable access to cartridge-based diagnostics.
QES: high confidence - For patients, access to clear, comprehensible and dependable information on what TB diagnostics are available to them and how to interpret results is a vital component of equity; lack of such access represents an important barrier for patients.
Interviews - New treatment options need to be matched with new diagnostics: it is important to improve access to treatment based on new diagnostics, and to improve access to diagnostics for new treatment options.
Interviews - The speed at which WHO guidelines are changing does not match the speed at which many country programmes are able to implement the guidelines. This translates into differential access to new TB diagnostics and treatment:
- between countries (i.e. between those that can and cannot quickly keep up with the rapidly changing TB diagnostic environment); and
- within countries (i.e. between patients who can and cannot afford the private health system that is better equipped to quickly adopt new diagnostics and policies).
Interviews
Is the intervention acceptable to key stakeholders?
- Acceptability of a high complexity reverse hybridization-based NAAT depends on how well the test performs on different samples, because laboratory staff question how well LPA methods work on smear-negative samples. If samples need to be cultured before the pyrazinamide LPA is run, this may undermine the benefits of this method’s quicker turnaround time compared with phenotypic DST for pyrazinamide. Acceptability also depends on how well the test actually detects mutations specific to pyrazinamide resistance; clinicians and laboratory staff may require further clarification and justification in some settings as to why this specific drug test is being prioritized, given that it is not currently part of routine DST.
- Specific feasibility challenges (training and infrastructure requirements, sample quality result interpretation system), general feasibility challenges (as identified in the interview study and QES, respectively) and accumulated delays risk undoing the added value and benefits identified by the users (e.g. avoiding delays and drug-resistance information).
QES high confidence and interviews
Is the intervention feasible to implement?
- The feasibility of implementing the pyrazinamide LPA is challenged by the significant training and laboratory infrastructure required to implement this method. Feasibility also hinges on the availability of an automated interpretation system, because the result is difficult to interpret.
Interviews
Implementation considerations
Factors to consider when implementing a high complexity hybridization-based NAAT for detection of pyrazinamide resistance are as follows:
- There are specific concerns about the complexity and difficulty of interpretation. The large number of bands makes it difficult to read the result of the high complexity reverse hybridization-based NAAT.
- Local epidemiological data on resistance prevalence should guide local testing algorithms, whereas pretest probability is important for the clinical interpretation of test results.
- The cost of a test varies, depending on the number of samples in a batch, staff time and other parameters requiring a local costing exercise to be performed.
- Low, moderate, and high complexity tests have a successive increase in technical competency needs (qualifications and skills) and staff time, impacting planning and budgeting.
- Availability and timeliness of local support service and maintenance should be considered when selecting a provider.
- Laboratory accreditation and compliance with a robust quality management system (including appropriate quality control) is essential for sustained service excellence and trust.
- Training of both laboratory and clinical staff will ensure effective delivery of services and clinical impact.
- Use of connectivity solutions for communication of results is encouraged, to improve efficiency of service delivery and time to treatment initiation.
- Based on a multinational, population-based study, levels of pyrazinamide resistance varied widely in the surveyed settings (3.0–42.1%). In all settings, pyrazinamide resistance was significantly associated with rifampicin resistance (49).
- Implementation of a high complexity hybridization-based NAAT requires laboratories with the required infrastructure, space and functional sample referral systems.
- Because there are several manual steps involved, well-trained staff are needed to set up assays and maintain instruments. Special training and experience are required for reading of banding patterns on the strip.
Research priorities
Research priorities for a high complexity hybridization-based NAAT for detection of pyrazinamide resistance are as follows:
- diagnostic accuracy of high complexity hybridization-based NAATs indirect testing on sputum and non-sputum samples in people with signs and symptoms of TB, with or without resistance to rifampicin;
- impact of diagnostic technologies on clinical decision-making and outcomes important to patients (e.g. cure, mortality, time to diagnosis and time to start treatment) in all patient populations;
- impact of specific mutations on treatment outcomes among people with DR-TB;
- use, integration and optimization of diagnostic technologies in the overall landscape of testing and care, as well as diagnostic pathways and algorithms;
- economic studies evaluating the costs, cost–effectiveness and cost–benefit of diagnostic technologies;
- qualitative studies evaluating equity, acceptability, feasibility and end-user values of diagnostic technologies; and
- interpretation of the results from a high complexity hybridization-based NAAT compared with sequencing and newer evidence on genotypic and phenotypic associations.
2.4.5 Targeted next-generation sequencing
Targeted NGS technology couples amplification of selected genes with NGS technology to detect resistance to many drugs with a single test. Also, since targeted NGS can interrogate entire genes to identify specific mutations associated with resistance, tests based on this technology may be more accurate than existing WRDs. In addition, new tests based on NGS can detect resistance to new and repurposed drugs that are not currently included in any other molecular assays. Hence, tests based on targeted NGS offer great potential to provide comprehensive resistance detection matched to modern treatment regimens.
Recommendations
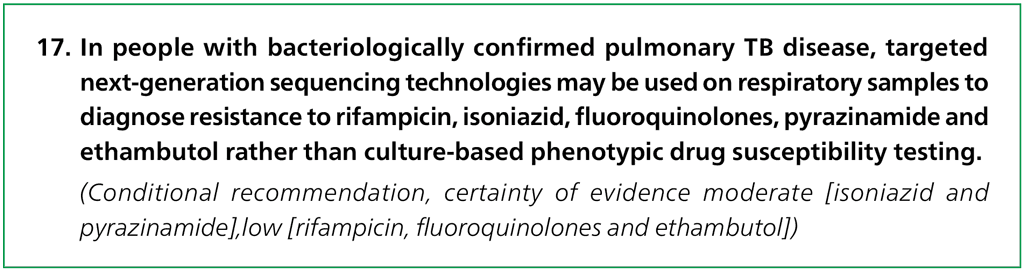
Remarks
- Priority should be assigned to those at higher risk of resistance to first-line treatment medications, including individuals who:
- continue to be smear or culture positive after 2 or more months of treatment, or experience treatment failure;
- have previously had TB treatment,
- are in contact with a person known to have resistance to TB drugs; or
- reside in settings or belong to subgroups where there is a high probability of resistance to either rifampicin, isoniazid or fluoroquinolone (used in new shorter regimens), or where there is a high prevalence of M. tuberculosis strains harbouring mutations not detected by other rapid molecular tests.
- This recommendation is conditional because of the lack of data on health benefits, the variable certainty of evidence on diagnostic accuracy, and the fact that accuracy is suboptimal for certain drugs. In addition, because this is a new technology that has not yet been widely implemented, there is still limited and variable evidence on costs, cost–effectiveness and feasibility of implementation.
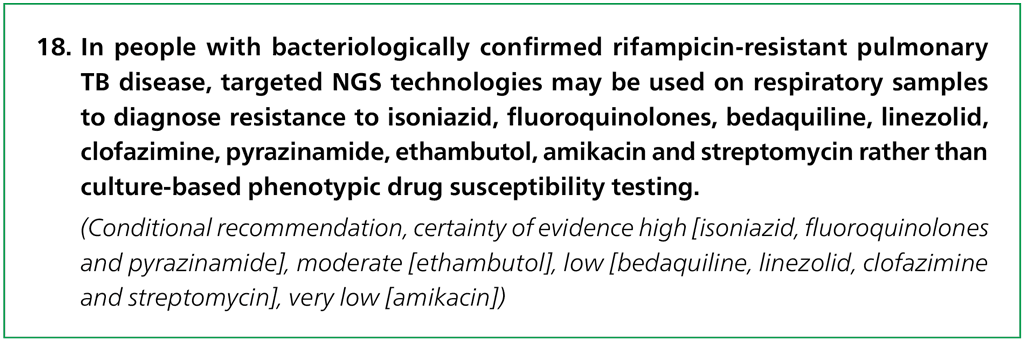
Remarks
- Priority should be given to those at a higher risk of resistance to medications used for the treatment of RR-TB, including individuals who:
- continue to be smear or culture positive after 2 months or more of treatment or have experienced treatment failure;
- have previously had TB treatment, including with the new and repurposed drugs;
- are in contact with a person known to have resistance to TB drugs, including the new and repurposed drugs; or
- have pre-XDR-TB with resistance to fluoroquinolones.
- As above, this recommendation is conditional because of the lack of data on health benefits, the variable certainty of evidence on diagnostic accuracy, the fact that accuracy is suboptimal for certain drugs, and limited and variable evidence on costs, cost–effectiveness and feasibility of implementation.

Test description
Three products met the inclusion criteria for detection of drug resistance to at least one of the anti-TB drugs under evaluation.
- The Deeplex® Myc-TB test (Genoscreen, France) is a targeted NGS-based kit for the simultaneous identification of mycobacterial species, genotyping and prediction of drug resistance of MTBC strains, directly applicable on sputum samples (50). The assay relies on deep sequencing of a 24-plex amplicon mix, and it targets 18 MTBC gene regions associated with resistance to anti-TB drugs (rifampicin, isoniazid, pyrazinamide, ethambutol, fluoroquinolones, amikacin, kanamycin, capreomycin, streptomycin, ethionamide, bedaquiline, clofazimine and linezolid). Mycobacterial species identification is performed by targeting the hsp65 gene; the spoligotyping target (CRISPR/Direct Repeat locus) and phylogenetic single nucleotide polymorphisms (SNPs) in targets associated with drug resistance are used for MTBC strain genotyping. The assay is performed using the Nextera XT and DNA Flex library preparation kits on the iSeq 100, MiniSeq, MiSeq and NextSeq sequencing platforms (Illumina). The solution includes an automated analysis pipeline of the sequencing data in a secure online application with integrated databases for results interpretation.
- The AmPORE-TB® test (Oxford Nanopore Diagnostics, United Kingdom) – previously referred to as Nano-TB) – is a targeted NGS-based kit for the simultaneous identification of mycobacterial species and the detection of MTBC genetic variants associated with antimicrobial resistance in DNA extracted from sputum samples.¹³ The assay relies on sequencing of a 27-plex amplicon mix: 24 drug-resistance targets, a genotyping target, a non-tuberculous mycobacteria (NTM) identification target (hsp65) and an internal control. The 24 drug-resistance targets are MTBC gene regions that are associated with resistance to various TB drugs (rifampicin, isoniazid, pyrazinamide, ethambutol, fluoroquinolones, amikacin, kanamycin, capreomycin, streptomycin, ethionamide, bedaquiline, clofazimine, linezolid and delamanid). Mycobacterial species identification is performed by targeting the hsp65 gene; the spoligotyping target (CRISPR/Direct Repeat locus) is used for MTBC strain genotyping. The assay is performed using the OND AmPORE-TB kit (OND-TBDR001-XX) and Flow Cells (OND-FLO-MIN001-XX) on the GridION Diagnostic Sequencing System (OND). The sequencing control software on the device can automatically start and report the results for the analysis workflows installed. The AmPORE-TB includes analysis software pre-installed on a device that processes readouts produced by the sequencing control software and creates an easy-to-interpret report, all performed locally on the device.
- The TBseq® test (Hangzhou ShengTing Medical Technology Co., China) is a kit based on targeted NGS that is used for the simultaneous identification of mycobacterial species and the prediction of drug resistance of MTBC strains; it is directly applicable to clinical specimens such as sputum and BAL fluid (51). The assay relies on deep sequencing of a multiplex amplification mix and it targets 21 MTBC genes associated with resistance to TB drugs (rifampicin, isoniazid, pyrazinamide, ethambutol, fluoroquinolones, amikacin, kanamycin, capreomycin, streptomycin, para-aminosalicylic acid, cycloserine, ethionamide or prothionamide, bedaquiline, clofazimine and linezolid). Mycobacterial species identification is performed by targeting the 16S and hsp65 gene regions. The assay is performed using the Universal Gene Sequencing Kit (ShengTing) to generate libraries that are sequenced on either a MinION or a GridION platform (Oxford Nanopore Technologies). The solution includes automated analysis software (Nano TNGS V1.0) for sequencing data processing and a secure online application (TBseq® Web App) with integrated databases for interpretation of results.
Justification and evidence
Diagnostic accuracy and health benefits
Two health questions were designed using the PICO approach, to form the basis for the evidence search, retrieval and analysis.
- Should targeted NGS as the initial test be used to diagnose drug resistance in individuals with bacteriologically confirmed pulmonary TB disease?
This question applies to:- rifampicin, using a composite reference standard of phenotypic DST and whole genome sequencing (WGS), and Xpert MTB/RIF® or Xpert Ultra® ;
- isoniazid, using phenotypic DST as the reference standard;
- levofloxacin, using phenotypic DST as the reference standard;
- moxifloxacin, using phenotypic DST as the reference standard;
- pyrazinamide, using a composite reference standard of phenotypic DST and WGS; and
- ethambutol, using a composite reference standard of phenotypic DST and WGS.
- Should targeted NGS be used to diagnose drug resistance in individuals with bacteriologically confirmed rifampicin-resistant pulmonary TB disease?
This question applies to:- isoniazid, using phenotypic DST as the reference standard;
- levofloxacin, using phenotypic DST as the reference standard;
- moxifloxacin, using phenotypic DST as the reference standard;
- pyrazinamide, using a composite reference standard of phenotypic DST and WGS;
- bedaquiline, using phenotypic DST as the reference standard;
- linezolid, using phenotypic DST as the reference standard;
- clofazimine, using phenotypic DST as the reference standard;
- amikacin, using phenotypic DST as the reference standard;
- ethambutol, using a composite reference standard of phenotypic DST and WGS; and
- streptomycin, using phenotypic DST as the reference standard.
A broad search was conducted to find, appraise and synthesize evidence about health benefits and the diagnostic test accuracy of targeted NGS compared with phenotypic drug sensitivity testing for patients with bacteriologically confirmed TB or with bacteriologically confirmed rifampicin-resistant pulmonary TB disease. A comprehensive search of three databases (Medline, Ovid Embase and Scopus) for relevant citations was performed. No date restriction was applied and the search was initially performed on 7 September 2022 and repeated on 17 January 2023. In addition, WHO made a public call for data and contacted well-known experts in the field to ask whether they had, or knew of, unpublished data that could contribute.
No data were found for the impact of targeted NGS on patient-level health effects. For the analysis of diagnostic accuracy, because few data were available in the literature, all data identified from the literature were included after correspondence with the authors. Hence, no manual data extraction from publications was required. A post-hoc decision was made to perform only an individual patient data (IPD) meta-analysis; thus, any study that could not provide IPD was excluded. Two report authors made independent assessments of methodological quality using QUADAS-2. Disagreements were resolved by discussion and uncertainties or disagreements were reviewed by an independent third party.
Subanalyses were performed to assess the diagnostic test accuracy in PLHIV and for semiquantitative results (derived from cycle thresholds) from Xpert MTB/RIF® or Xpert Ultra® , where “very low” or “low” concentrations of M. tuberculosis were compared with “medium” or “high” concentrations. The very low or low semiquantitative categories represent paucibacillary disease states, such as those frequently observed in paediatric TB.
Data were included from both published and unpublished prospective, observational clinical studies of targeted NGS platform diagnostic accuracy. All studies where targeted NGS had been performed directly from processed clinical samples were included, whereas those performed exclusively on cultured isolates were excluded. All studies were required to have comparator phenotypic DST data as a reference; in the cases of rifampicin, ethambutol and pyrazinamide, studies were required to also have WGS, to allow a composite reference to be generated. Rifampicin resistance results and semiquantitative results from Xpert MTB/RIF® or Xpert Ultra® were requested from all studies.
Given that this was a review of the diagnostic accuracy of a class of diagnostic platforms, all the data from each platform alone were analysed to assess which to include in an analysis to inform a class recommendation. Where the performance of any one platform appeared to be an outlier for sensitivity or specificity, that platform was excluded from subsequent metaanalyses. A platform was considered to be an outlier for a particular drug if the point estimate for sensitivity was more than 10 percentage points worse than the best performing platform, or where the point estimate for specificity was more than 5 percentage points worse.
An IPD meta-analysis was performed instead of a classical meta-analysis, because the studies identified in the literature were generally too small to contribute to a classical meta-analysis, particularly for the new and repurposed drugs. In addition, this type of approach allowed for relevant co-variables to be included in the model; it could also control for repeated testing on the same samples using different platforms, which was the case for much of the available data.
For each dependent variable, a multivariable model included a number of co-variables as fixed effects. These included rifampicin resistance as determined by Xpert MTB/RIF® or Xpert Ultra® for all drugs other than rifampicin; semiquantitative cycle threshold (CT) value from Xpert MTB/RIF® or Xpert Ultra® ; and a co-variable to indicate which samples featured in duplicate, meaning that some samples were sequenced on two different platforms and thus were represented twice in the analysis. For models looking specifically at diagnostic test accuracy in PLHIV, the HIV test result was included as a co-variable. Finally, the study site was included as a random effect. The models were run in Stata (version 17) using the melogit command, and the outputs were transformed using the margins command. Models were run for all PICO questions for sensitivity and specificity.
The certainty of the evidence of the pooled studies was assessed systematically for each of the PICO questions using the GRADE approach, which produces an overall quality assessment (or certainty) of evidence and has a framework for translating evidence into recommendations.
The GRADEpro Guideline Development Tool software (16) was used to generate summary of findings tables for the sensitivity and specificity of each drug. The numbers of samples classified as true, false positive or negative were then calculated across a range of three prevalences of drug resistance, chosen to be representative of different global settings. The quality of evidence was rated as high (not downgraded), moderate (downgraded one level), low (downgraded two levels) or very low (downgraded more than two levels), based on five factors: risk of bias, indirectness, inconsistency, imprecision and other considerations. The quality (certainty) of evidence was downgraded by one level when a serious issue was identified and by two levels when a very serious issue was identified in any of the factors used to judge the quality of evidence.
The data sources for the IPD data analysis are shown in Fig. 2.4.5.1. The analysis included data from published studies, a large multicountry trial conducted by FIND, and several other studies across multiple countries. Most of the studies only evaluated the Deeplex assay, while the FIND trial evaluated both the Deeplex and the AmPORE-TB. Only one study evaluated TBseq. For each drug, one or two platforms were dropped from the analysis based on the overall number of resistant or susceptible samples available for that platform and drug, or because the accuracy of the platform did not meet the diagnostic test accuracy criteria for inclusion when compared with the best performing platform.
Fig. 2.4.5.1 Studies included in the IPD meta-analysis for targeted NGS
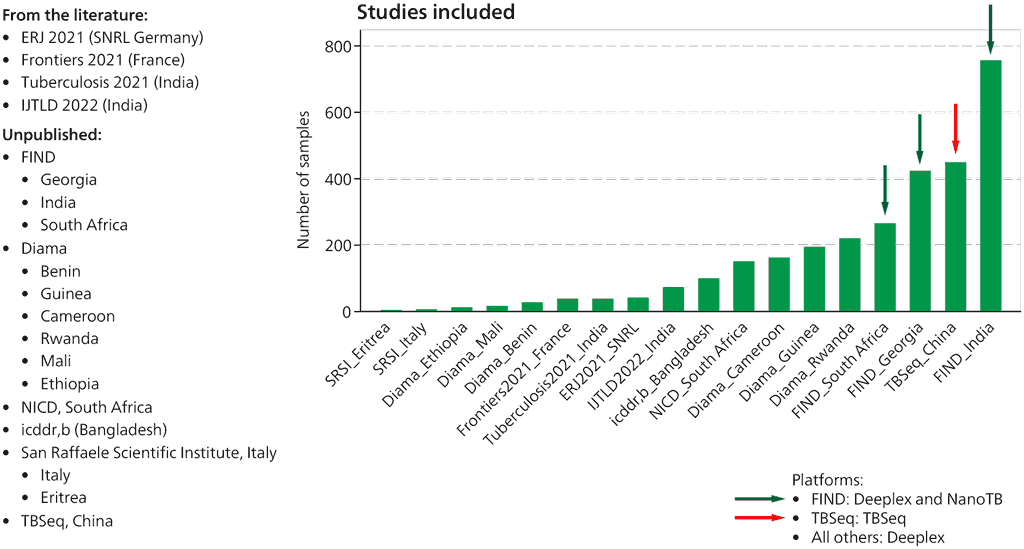
ERJ: European Respiratory Journal; FIND: Foundation for Innovative New Diagnostics; IPD: individual patient data; NGS: next-generation sequencing; NICD: National Institute for Communicable Diseases; UTLD: International Union Against Tuberculosis and Lung Diseases.
Data synthesis was structured around the two preset PICO questions, as outlined below.
PICO 1: Should targeted NGS as the initial test be used to diagnose drug resistance in patients with bacteriologically confirmed pulmonary TB disease?
The available evidence included in the final pooled analysis varied by drug, from 12 studies with 1440 participants for the sensitivity of isoniazid to three studies with 269 participants for the specificity of pyrazinamide (Table 2.4.5.1). The pooled estimates were determined using a multivariable, mixed-effects model. All drugs were downgraded by one level for indirectness for sensitivity and specificity, because all studies were enriched for rifampicin resistance, leading to applicability concerns. In addition, for rifampicin, levofloxacin and pyrazinamide, specificity was downgraded a further level for imprecision; however, for ethambutol, it was downgraded for risk of bias because different samples were used for the index and reference tests. The overall certainty of the evidence for test accuracy ranged from moderate to very low.
The test performance was determined to be accurate for all drugs included in the assessment, with a pooled sensitivity of at least 95% for isoniazid, moxifloxacin and ethambutol, more than 93% for rifampicin and levofloxacin, and 88% for pyrazinamide. The pooled specificity was at least 96% for all drugs.
The reference standard was culture-based phenotypic DST for isoniazid, levofloxacin and moxifloxacin, and a combination of phenotypic DST and WGS for rifampicin, pyrazinamide and ethambutol. The percentage of tests with indeterminate results ranged from 9% (levofloxacin and moxifloxacin) to 18% (pyrazinamide), with higher indeterminate rates in samples with lower bacterial load (semiquantitative category low or very low).
Table 2.4.5.1 The accuracy and certainty of evidence of targeted NGS for the detection of resistance to anti-TB drugs among bacteriologically confirmed pulmonary TB
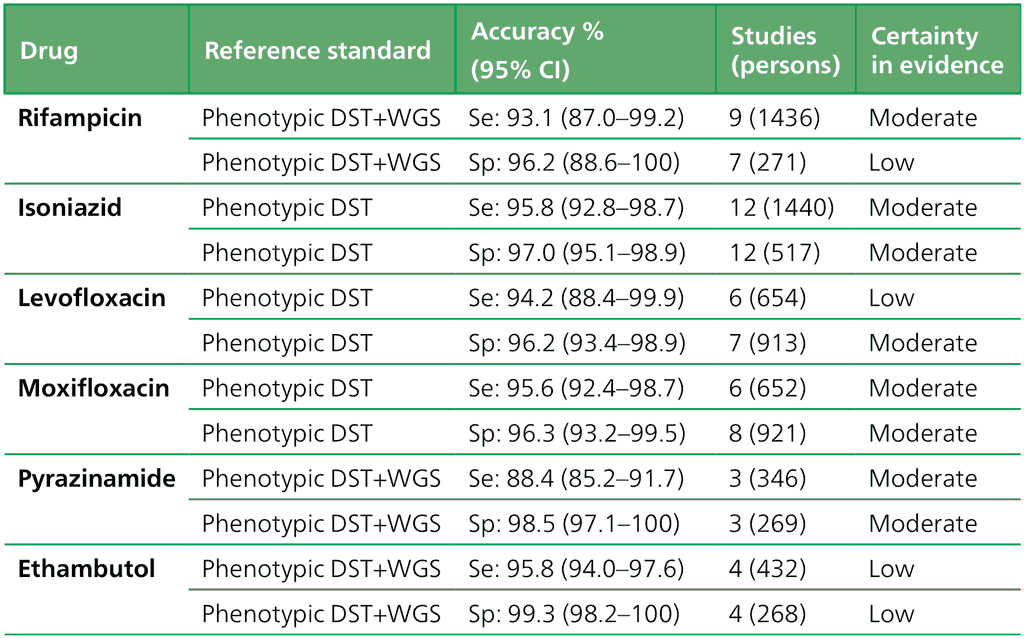
CI: confidence interval; DST: drug susceptibility testing; NGS: next-generation sequencing; Se: sensitivity; Sp: specificity; TB: tuberculosis; WGS: whole genome sequencing.
There were no data on the impact of targeted NGS on patient outcomes such as time to treatment or treatment outcome.
PICO 2: Should targeted NGS be used to diagnose drug resistance in patients with bacteriologically confirmed rifampicin-resistant pulmonary TB disease?
The available evidence varied by drug, from 12 studies with 1440 participants for sensitivity of isoniazid to three studies with 31 participants for sensitivity of bedaquiline (Table 2.4.5.2). The pooled estimates were determined using a multivariable, mixed-effects model.
The overall certainty was high for some of the drugs. Levofloxacin was downgraded one level for inconsistency. Bedaquiline and linezolid were downgraded by two levels for imprecision in sensitivity because the number of resistant samples was below the threshold set and the confidence intervals were wide. Clofazimine was also downgraded by two levels, one for inconsistency (because two studies were outliers) and another level for imprecision (because the confidence intervals were wide). Amikacin was downgraded by one level for sensitivity and specificity because critical concentrations outside those recommended by WHO were used for a large proportion of samples. Amikacin sensitivity was further downgraded by two more levels, one for inconsistency and the other for imprecision. Ethambutol was downgraded by one level for risk of bias because different samples were used for the index and reference tests. Streptomycin specificity was downgraded by two levels, one for inconsistency and the other for imprecision. The overall certainty of the evidence for test accuracy ranged from high to very low.
The test performance among people with RR-TB was determined to be accurate for isoniazid, levofloxacin, moxifloxacin, ethambutol and streptomycin (pooled sensitivity ≥95%) and acceptable for pyrazinamide (90%), bedaquiline (68%), linezolid (69%), clofazimine (70%) and amikacin (87%). The pooled specificity was 95% or greater for all drugs except streptomycin (75%). The reference standard was culture-based phenotypic DST for all drugs except for ethambutol and pyrazinamide, where a combination of phenotypic DST and WGS was used. The percentage of tests with indeterminate results ranged from 9% (levofloxacin and moxifloxacin) to 21% (ethambutol); indeterminate rates were higher in samples with a lower bacterial load (semiquantitative category low or very low).
Table 2.4.5.2 The accuracy and certainty of evidence of targeted NGS for the detection of resistance to anti-TB drugs among bacteriologically confirmed rifampicin-resistant pulmonary TB
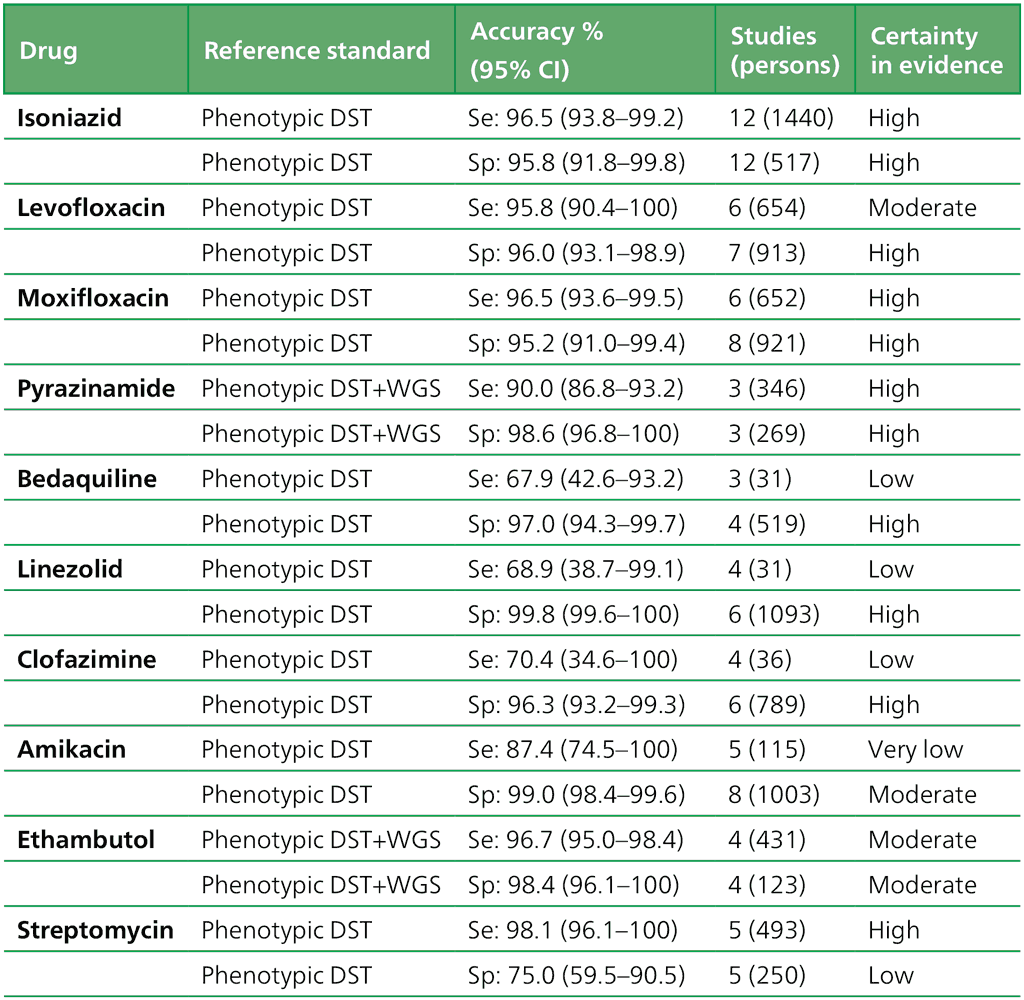
CI: confidence interval; DST: drug susceptibility testing; NGS: next-generation sequencing; Se: sensitivity; Sp: specificity; TB: tuberculosis; WGS: whole genome sequencing.
There were no data on the impact of targeted NGS on patient outcomes such as time to treatment or treatment outcome.
Three web annexes give additional information, as follows:
- details of studies included in the current analysis (Web Annex A.10: Review of the diagnostic accuracy of targeted NGS technologies for detection of drug resistance among people diagnosed with TB);
- a summary of the results and details of the evidence quality assessment (Web Annex A.10: GRADE profiles of targeted next-generation sequencing for detection of TB drug resistance); and
- a summary of the GDG panel judgements (Web Annex A.10: Evidence to decision tables: targeted next-generation sequencing for detection of TB drug resistance).
Cost–effectiveness analysis
The cost and cost–effectiveness data for targeted NGS were assessed through a systematic review of the published literature and a generalized model-based cost–effectiveness analysis commissioned by WHO.
The systematic review on the cost and cost–effectiveness of using either targeted NGS or WGS to diagnose DR-TB searched three databases: PubMed, Embase and Scopus. The search was run on 30 October 2022 and had no time restriction. All costing data were inflated to 2021 US dollars. Findings were synthesized descriptively, given the considerable degree of heterogeneity in study methodology and outcomes. Among the studies included in the systematic review, three were on targeted NGS only, three were on targeted NGS and WGS, and four were on WGS only. For targeted NGS based on a single study (n=1), the cost per sample was between US$ 69.64 for Illumina MiSeq on 24 samples, and US$ 73.47 for Nanopore MinION on 12 samples; however, this costing was limited to only some components and did not include human resource costs or overhead costs. For WGS (n=5), cost per sample ranged from US$ 63.00 on Nanopore MinION to US$ 277.00 on Illumina MiSeq; given that studies used an inconsistent number of component costs, comparisons were challenging. Based on the review, the most significant cost component was the sequencing step, and the largest component costs were reagents and consumables, including those necessary for sequencing, sample processing and targeted NGS steps library preparation. Study authors identified four major cost drivers: use of different sequencers, depth and breadth of coverage, inefficiencies in initial sample runs, and economies of scale via batching or cross-batching.
The cost data from the systematic review were limited; therefore, an empirical unit costing was performed, in consultation with manufacturers and FIND. At the time of this work, only pricing for Deeplex Myc-TB was available and it was used for estimation of cost for the class. Unit costs included consumables, equipment, staffing and overheads (where available); also, costs assumed targeted NGS testing for all drugs. Based on the empirical analysis, the cost of targeted NGS was estimated to be:
- US$ 134 to US$ 257 in South Africa;
- US$ 120 to US$ 198 in Georgia; and
- US$ 121 to US$ 175 in India.
These costs are dependent on patient volume, batching and negotiated cost per targeted NGS kit.
Recognizing the lack of economic evidence on this topic, a hypothetical cost–effectiveness modelling study was undertaken to assess the cost–effectiveness (Objective 1) and affordability (Objective 2) of these tests for the diagnosis of DR-TB in various high TB burden settings.
Objective 1: To assess the potential cost–effectiveness of introducing the targeted NGS technology for the diagnosis of DR-TB in Georgia, India and South Africa.
This assessment included modelling the cost–effectiveness of targeted NGS in three separate scenarios with distinct comparison options:
- Cost–effectiveness of targeted NGS for DST among individuals with RR-TB after a rapid molecular test for rifampicin resistance as a replacement for phenotypic DST (PICO 2).
- Cost–effectiveness of targeted NGS for DST among individuals with RR-TB after a rapid molecular test for rifampicin resistance as a replacement for current in-country DST practice (PICO 2).
- Cost–effectiveness of targeted NGS as the initial test for TB drug resistance in patients with bacteriologically confirmed TB compared with rapid molecular testing for drug resistance and phenotypic DST in a high DR-TB burden setting (PICO 1).
In the first scenario, targeted NGS was compared with universal phenotypic DST; in the second scenario, targeted NGS was compared with current in-country phenotypic DST practice among individuals with detected rifampicin resistance (PICO 2). This was done across three countries: Georgia, India and South Africa. Current DST practice in Georgia and South Africa includes Xpert XDR® followed by phenotypic DST; in India it includes LPAs and phenotypic DST done in parallel. A final scenario included targeted NGS compared to rapid molecular testing for drug resistance and phenotypic DST as initial tests for TB drug resistance among all TB patients (PICO 1) but was modelled for only one setting, Georgia – a high DR-TB burden setting. Epidemiological data were sourced from published literature; targeted NGS diagnostic accuracy data were sourced from the systematic review and IDP analysis conducted for this guideline. Economic data were sourced from published literature and a systematic and scoping review done in parallel by our team and supplemented with empirical data collection.
A decision analysis modelling approach was used to estimate the incremental cost–effectiveness of using targeted NGS for the diagnosis of DR-TB compared with various existing DST scenarios. This was done from the perspective of the health care system and accounts only for the health care system costs required to diagnose and treat TB. The estimation did not account for societal costs, or any direct or indirect costs incurred by patients. In addition, costs for sample transportation were not included in this analysis. The primary outcome was the incremental cost–effectiveness ratio (ICER), which was calculated as the incremental cost in US dollars per disability-adjusted life year (DALY) averted.
Main findings for PICO 1: Using targeted NGS as an initial test
Using targeted NGS as an initial test for DST in the high DR-TB burden setting of Georgia led to more health gains (DALYs=0.49) compared with Xpert MTB/RIF or Xpert Ultra, followed by phenotypic DST (DALY=0.51). The ICER per DALY averted was US$ 9261 (95% uncertainty range [UR]: US$ 5258–32 040/DALY averted), which was considered cost effective at a willingnessto-pay (WTP) threshold of three times the country GDP per capita (US$ 15 609), with 80% of simulated iterations falling below the WTP threshold.
Main findings for PICO 2: Using targeted NGS among those with RR-TB
Using targeted NGS as a replacement for universal phenotypic DST among RR-TB patients, targeted NGS was dominated by phenotypic DST, with targeted NGS having higher costs and leading to fewer health gains. This finding was driven by the high diagnostic accuracy of phenotypic DST (which was assumed to be universal in this scenario), and an assumption of no difference in loss to follow-up between targeted NGS and phenotypic DST. When in-country DST practice was used as the comparator (instead of universal phenotypic DST), targeted NGS led to more health gains than in-country DST across all three countries. Targeted NGS was cost effective in South Africa (ICER: US$ 15 619/DALY averted, 95% UR: cost saving –US$ 114 782, at a WTP threshold of US$ 21 165), but was not cost effective in Georgia (ICER: US$ 18,375/ DALY averted, UR: cost saving –US$ 158 972/DALY averted, at a WTP threshold of US$ 15 065). In India, where LPA, liquid culture and DST are being used as part of in-country DST, targeted NGS dominated the country’s current DST practice, with lower costs and more health gains (95% UR: cost saving –US$ 60 083).
Main findings: scenario analyses
Several key scenario analyses were investigated. In the base case approach, loss to follow-up was assumed to be equivalent between phenotypic DST and targeted NGS; in a scenario where there was no loss to follow-up in targeted NGS compared with 10% in phenotypic DST, targeted NGS was cost effective in South Africa (ICER: US$ 13 004/DALY averted, WTP: US$ 21 165) and Georgia (ICER: US$ 13 640/DALY averted, WTP: US$ 15 069) and targeted NGS still dominated in-country DST practice in India. In scenarios where sequencing platforms are used for multiple different diseases to reduce the unit test cost of targeted NGS, the cost–effectiveness of targeted NGS improves in all three countries. A batching scenario was investigated, with an assumed 20% fewer samples per targeted NGS run, and led to an increased unit test cost for targeted NGS; in this scenario, the targeted NGS approach retained cost–effectiveness only in South Africa. When a 50% price reduction in targeted NGS test kit cost was assumed, targeted NGS cost–effectiveness further improved in all countries.
Objective 2: To assess the financial impact of introducing targeted NGS as a replacement for existing DST for diagnosis of DR-TB among TB patients across three countries: Georgia, India and South Africa.
A budget impact assessment was undertaken to estimate the financial consequences of adopting targeted NGS for DST for all patients diagnosed with TB, and replacing in-country DST practice in Georgia (PICO 1). The analysis suggested that implementing targeted NGS for all patients diagnosed with TB would be more expensive than testing all patients with Xpert MTB/RIF or Xpert Ultra, followed by phenotypic DST (see Fig. 2.4.5.2).
Fig. 2.4.5.2 Budget impact assessment results comparing current standard practice for DST with implementation of targeted NGS for all patients diagnosed with TB in Georgia
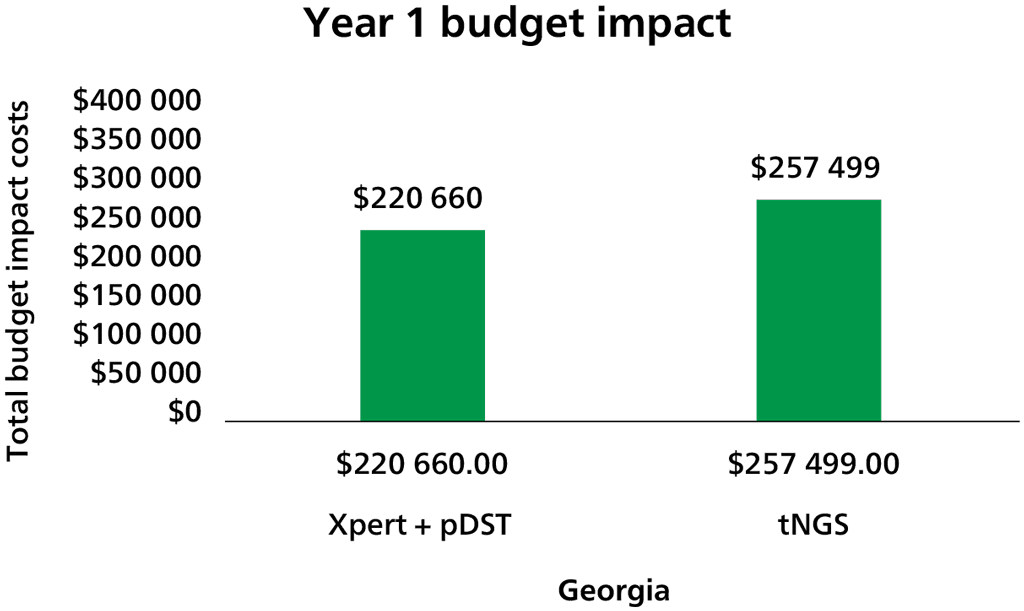
DST: drug susceptibility testing; NGS: next-generation sequencing; pDST: phenotypic DST; TB: tuberculosis; tNGS: targeted NGS.
A budget impact assessment was undertaken to estimate the financial consequences of adopting targeted NGS for DST after a rapid molecular test for rifampicin resistance, and replacing in-country DST practice in Georgia, India and South Africa (PICO 2). In-country DST practice included Xpert XDR combined with phenotypic DST in Georgia and South Africa, and Xpert XDR combined with LPA in Georgia over a 1-year and 5-year period. It was assumed that the eligible RR-TB patient populations requiring DST were 58 837, 8200 and 187 in South Africa, India and Georgia, respectively, and that the TB reduction rate over the 5 years was stable (2). To estimate the impact on the country-specific budget, the economic costs generated by the model were multiplied by the number of patients.
Results from a 1-year budget impact assessment for PICO 2 are presented in Fig. 2.4.5.3 In India, it was estimated that implementing targeted NGS would cost about US$ 57 130 727 – slightly lower than the current practice of LPA combined with phenotypic DST, which has a cost of US$ 57 719 097. In South Africa, it was estimated that implementing targeted NGS would result in a rise in budget to about US$ 27 888 200, slightly more than LPA combined with phenotypic DST, which has a cost of US$ 26 428 600. Finally in Georgia, where there are fewer bacteriologically confirmed patients, it was estimated that implementing targeted NGS would cost about US$ 592 221, slightly more than LPA combined with phenotypic DST, which has a cost of US$ 568 480.
Fig. 2.4.5.3 Budget impact assessment results comparing current standard practice for DST to implementing targeted NGS for patients with RR-TB in India, South Africa and Georgia
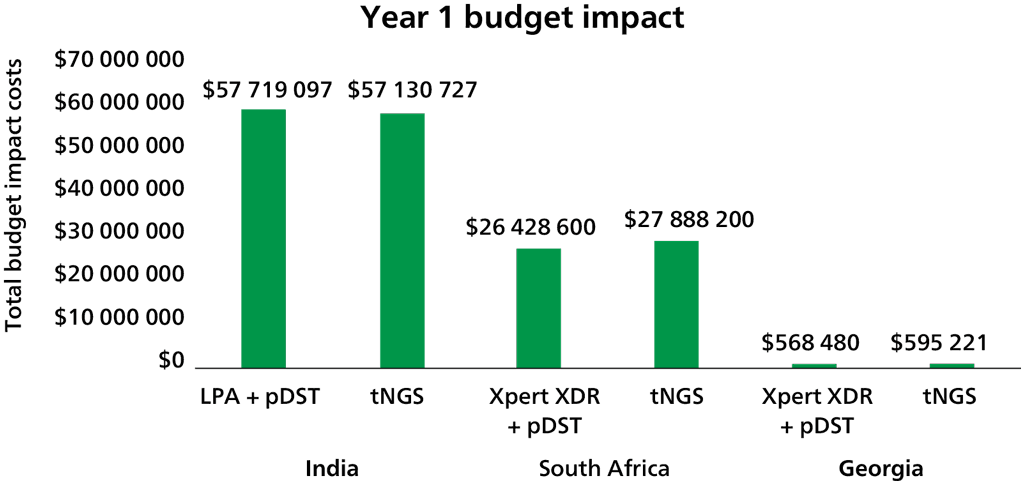
DST: drug susceptibility testing; LPA: line probe assay; NGS: next-generation sequencing; pDST: phenotypic DST; RR-TB: rifampicin-resistant TB; TB: tuberculosis; tNGS: targeted NGS.
User perspective
A rapid review was commissioned to identify and synthesize qualitative evidence on the use of targeted NGS for the detection of TB drug resistance; in particular, the aim was to examine the implementation considerations related to acceptability, feasibility, and values, preferences and equity. The review searched Medline with no year or language limits. The search was run on 19 August 2022, and then rerun on 10 October 2022 to include WGS-related studies for the detection of TB drug resistance. The review did not identify any eligible studies for analysis and synthesis. Based on the systematic search, three records were identified; in addition, based on the open, hand and expert searches, 27 records were found. On full-text review of the 30 records, none were found to be eligible for inclusion. Given that no direct evidence was found, note was made of a Cochrane qualitative evidence synthesis published in 2022 that examined recipient and provider perspectives on rapid molecular tests for TB and drug resistance (52); that study provides relevant (though indirect) evidence on the subject. The authors noted that people with TB valued reaching diagnostic closure with an accurate diagnosis, avoiding diagnostic delays and keeping diagnostic associated costs low, whereas health care providers valued aspects of accuracy and the resulting confidence in low complexity NAAT results, rapid turnaround times and low costs to people seeking a diagnosis.
To address the direct evidence gap, WHO commissioned an additional qualitative cross-sectional study comprising semi-structured interviews, primarily with laboratory staff and management personnel directly involved with implementing targeted NGS in the three FIND trial sites, as well as with three global experts involved in TB care and diagnostics. In total, there were 17 respondents, and the work was conducted during September to October 2022. The objective was to explore the perceptions and experiences of those implementing targeted NGS technology, with respect to acceptability, feasibility, and values, preferences and equity. The main findings are summarized below.
Acceptability
A consistently positive sentiment was expressed for the acceptability and potential utility of targeted NGS technology. Targeted NGS was seen as a “major advancement” in molecular MDR-TB diagnostics.
- The main reasons for the high level of acceptability were the comprehensiveness (resistance diagnosis for more drugs and for the newest and repurposed drugs), the convenience of using a sputum sample (as compared with culture samples), and the rapidity (quick results compared with phenotypic testing times; 3–5 days as compared with 4–6 weeks).
- There was also the sense that there is a good window of opportunity to benefit from the utility of targeted NGS technology; that is, the technology is arriving at the right time, given that resistance to newer TB drugs is likely to increase as the use of these drugs becomes routine.
Feasibility
Although there was high praise for the capability and potential utility of targeted NGS technology, several challenges were identified when testing samples using the targeted NGS platforms, which may limit the feasibility of targeted NGS for routine uptake at the present time. The overall sentiment was that the targeted NGS technology needs to be further developed before it can be considered fully ready for operational use.
The following feasibility challenges were identified:
- Start-up and setting-up challenges: Multiple problems were identified with starting and setting up the technology. These problems related to the newness of the technology and the trial setting, importing technology and specialist supplies, lack of in-country technical assistance for problem-solving and need for more hands-on training practice.
- High technical complexity of the test: Targeted NGS technology was seen as a high complexity molecular test that was technically challenging. For example, preparing the sample for sequencing involves multiple steps that require attention to detail and precision, leaving little room for error. Preparation of the library is particularly complex for the Deeplex platform, although both the Deeplex and the Nanopore platforms are quite complex. In both platforms, it was thought that there were too few opportunities for early recognition and correction of errors, increasing the risk of failed runs.
- Specialized laboratory infrastructure and human resource requirements: Because targeted NGS is a molecular-based testing platform, it requires highly specialized laboratory infrastructure (e.g. multiple rooms to prevent amplicon contamination and specialized cold storage facilities). Also, highly specialized molecular and medical scientists are needed to perform the tests. In LMIC settings, such specialized laboratory infrastructure and staff may only be available at centralized laboratories (i.e. not at regional laboratories).
- Special requirements for operating the test: In addition to highly specialized laboratory infrastructure and staff, the testing technology also requires an uninterrupted supply of electricity, high internet connectivity, high computer capacity, clean water and temperature controls – requirements that may pose challenges in some LMIC settings.
- Supply chain challenges: Major challenges were reported relating to the required supply chain for implementing targeted NGS. Procurement bottlenecks and delays coupled with shelf-life limitations of reagents jeopardize continuous access to specialist supplies.
- Data management and storage requirements: There were concerns that data analysis and data storage requirements were not fully developed, including systems for backing up data, ownership of data and security of data. Another issue that needs to be considered is how targeted NGS and routine laboratory information systems can be interlinked.
- Continuous updating of the WHO catalogue of mutations is required: There was agreement that the usefulness of the targeted NGS technology depends on the informational support provided by the WHO catalogue of mutations (53), which allows for meaningful interpretation of resistance data; thus, there is a need for the WHO catalogue to be continuously updated.
- Feasibility concerns differed for the different targeted NGS platforms: The overall sentiment was that all targeted NGS platforms needed to be further developed before they are fully ready for operational use, some more than others. The high level of technical complexity of the sample preparation stages (mainly the library preparation stage) was considered a key challenge for the Deeplex platform, and the need for improved computer analysis and storage capacity was a challenge for the Oxford Nanopore platform, although both required a high level of precision and attention to detail. There is also a need to incorporate steps for early error recognition.
Values, preferences and equity
The overall sentiment is that MDR-TB diagnostic technology needs to balance accuracy, speed, affordability, equity and cost–effectiveness, and that targeted NGS technology would need to address these considerations before it can be implemented in LMIC settings. These considerations were consistent across the different stakeholder groups who participated in the study.
- Centralized versus decentralized placement may have equity implications for access: Given the high-level specialized laboratory infrastructure, specialized human resources and technical complexity needed for targeted NGS, the technology may be suitable for placement only at centralized, reference laboratories. This may have equity access considerations if it means less access for some regions of a country that lack reference laboratories. This may also have implications for costs (e.g. costs for transport of sputum), probability of sample loss and time to results.
- Affordability and cost–effectiveness are major concerns: There was a major concern about the financial costs of the targeted NGS technology and the affordability for LMIC. Participants were worried about the cost of the equipment and the costs of ongoing specialist supplies (especially reagents), as well as the cost of maintaining equipment. They noted that costing calculations should be comprehensive and should include the cost of special consumables, extra general laboratory consumables and additional infrastructure needs (e.g. extra space, temperature control and internet connectivity). There were concerns that cost–effectiveness calculations should be comprehensive and should include assessment of the impact of the use of targeted NGS testing on improving TB outcomes.
- The MDR/RR-TB case burden of a country could influence equitable access at centralized levels. In some settings with high caseloads, the targeted NGS technology capacity in central laboratories may not be sufficient for processing large caseloads in good time; also, in settings with low caseloads, waiting for sufficient samples to batch-test will cause delays.
Implementation considerations
Although the evidence that is available supports the use of targeted NGS to detect drug resistance after TB diagnosis, to guide clinical decision-making for DR-TB treatment, the following factors need to be considered when implementing these tests:
- Regulatory approval from national regulatory authorities or other relevant bodies is required before implementation of these diagnostic tests.
- In its current format, targeted NGS is a high complexity test that is most suitable for centralized laboratories equipped with specialized skills and infrastructure.
- Targeted NGS tests do not replace existing rapid tests that are more accessible and easier to perform for detecting resistance to rifampicin, isoniazid and fluoroquinolones. However, if targeted NGS can be performed rapidly, it can be considered as an alternative initial option for prioritized populations. Those who will benefit most from these tests are individuals who require rapid and comprehensive DST but have limited access to phenotypic DST.
- Priority should be given to samples with a high bacillary load as determined by initial bacteriological tests (e.g. semiquantitative high/medium or smear-positive grading). In situations where the bacillary load is low (e.g. semiquantitative low/very low/trace or smear-negative grading), the recommendations still hold, although rates of indeterminate results are likely to be higher; therefore, phenotypic DST is likely still required for samples with a low bacillary load.
- Similarly, the recommendations apply to children, adolescents and PLHIV populations because these populations have a higher frequency of samples with low bacterial load.
- The recommendation is based on data obtained from sputum and BAL specimens, and can be extrapolated to other lower respiratory tract samples (e.g. endotracheal aspirates). However, further research is needed to evaluate the use of these tests on alternative sample types for diagnosing pulmonary TB in children (e.g. nasopharyngeal and stool samples) and diagnosing extrapulmonary TB.
- Since sensitivity for bedaquiline, linezolid and clofazimine resistance is suboptimal, consideration of the pretest probability is important in interpreting the targeted NGS results for these drugs. Further testing of samples with a susceptible result (using culture-based phenotypic DST) would be warranted, particularly when the risk of resistance is high. Since specificity is high, a result that indicates resistance may be used to guide the therapy, particularly among those at risk for resistance. In the case of pretomanid, the basis for resistance has not been fully elucidated; hence, culture-based DST is also required for this drug.
Research priorities
Several key research priorities emerged from the reviews of the available evidence on targeted NGS for detecting TB drug resistance. They fall into three main categories: clinical research, implementation research, and monitoring and evaluation.
Clinical research:
- Conduct clinical trials to assess the impact of targeted NGS on patient-important outcomes¹⁴.
- Evaluate the accuracy and impact on patient-important outcomes of targeted NGS among populations of individuals diagnosed with TB, across a range of prevalences of rifampicin or other drug resistance).
- Assess the accuracy and impact on patient-important outcomes of targeted NGS for detecting resistance to new and repurposed drugs, including pretomanid, across varied geographical and epidemiological settings.
- Assess the accuracy and impact on patient-important outcomes of targeted NGS for analysing extrapulmonary samples, including CSF for meningitis, non-sputum samples (e.g. nasopharyngeal aspirate, gastric aspirate or stool) for children, and alternative sample types (e.g. tongue swabs) in both adults and children.
- Undertake additional qualitative and quantitative research to further understand the perspectives of end-users and clinicians regarding the acceptability and feasibility of using targeted NGS.
Implementation research:
- Develop and evaluate effective and efficient implementation models by integrating targeted NGS into laboratory networks and optimizing algorithms, with the aim of enhancing timely access to testing and treatment initiation, and improving patient outcomes.
- Develop strategies to enhance the efficiency of targeted NGS testing, including sample processing and concentration techniques, determining optimal thresholds of bacterial load from initial tests before performing targeted NGS, and employing molecular transport medium for the ambient storage and transfer of samples to testing sites.
- Regularly update the WHO catalogue of mutations (53), incorporating additional genetic targets and including new drugs (e.g. pretomanid) to enhance the sensitivity and specificity of targeted NGS.
- Explore technological advancements to simplify the testing process, automate steps (especially library preparation), develop decentralized targeted NGS solutions and investigate potential synergies with existing initial tests (e.g. using leftover DNA or smear-positive slides).
- Conduct comprehensive mapping of sequencing capacity within countries and perform diagnostic network optimization exercises. Placement of the technology should consider the demand for sequencing across multiple diseases, facilitating cross-disciplinary use of the machines and shared costs.
- Compile and use lessons learned from applying targeted NGS technology in other diseases (e.g. COVID-19) to develop effective implementation strategies for TB.
Monitoring and evaluation:
- Standardize the nomenclature for reporting of results across different targeted NGS technologies, for integration into health information data systems.
- Ensure separate recording of true failures and unclassified mutations, and monitor trends over time as an essential component of result reporting.
- Regularly monitor performance data, including overall resistance rates, resistance rates by specific drugs or targets and turnaround times (both total and in-laboratory).
- Incorporate quality monitoring measures, such as tracking indeterminate rates, sequencing coverage and depth, and participating in external quality assurance programmes.
- Establish an external quality assurance programme for sequencing that covers all relevant targets of interest.
- Integrate the sequencing data generated into existing surveillance systems to monitor the prevalence and trends in drug resistance effectively. Share the data to update the WHO mutation catalogue.
- Collect cost data to address important questions, such as the costs associated with introducing and scaling up targeted NGS in different settings, the trade-offs between turnaround time and batching, and the optimal balance in various settings.
- Assess the impact of multidisease testing on programme operations and costs, including disease-specific testing volumes, turnaround times, costing, resource sharing and resource requirements.
- Evaluate the impact of time to treatment initiation or modification, treatment outcomes and overall cost–effectiveness of targeted NGS implementation.
12 Leen Rigouts: Validation study of Genoscholar PZA LPA in three Supranational TB Reference Laboratories.
13 Oxford Nanopore Diagnostics provided a draft protocol for the test.
14 Mortality, Cure, Lost to follow up; Time to diagnosis; Time to treatment.

 Feedback
Feedback